Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
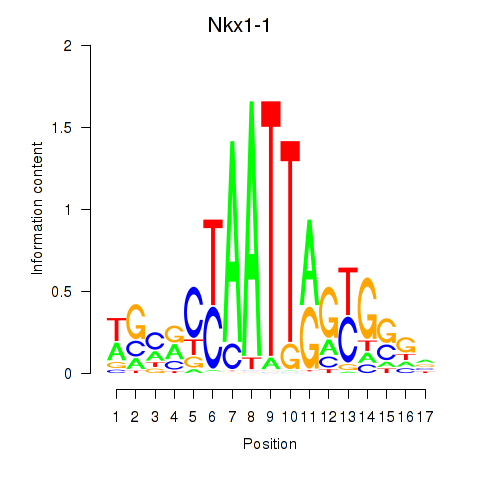
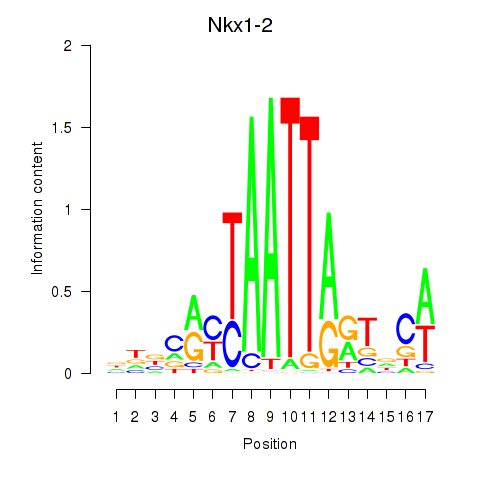
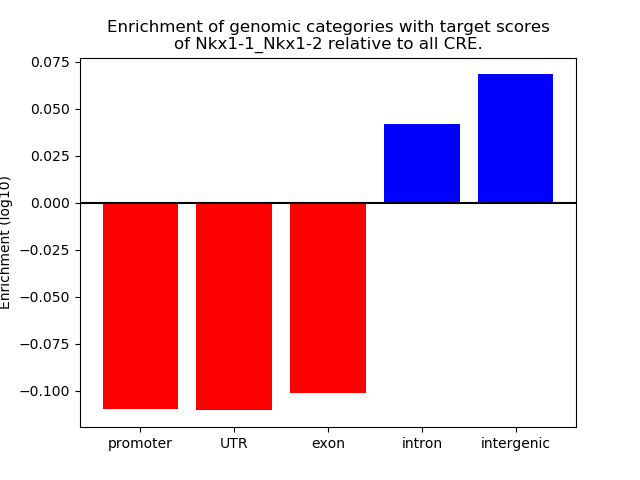
Results for Nkx1-1_Nkx1-2
Z-value: 0.90
Transcription factors associated with Nkx1-1_Nkx1-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx1-1
|
ENSMUSG00000029112.5 | NK1 homeobox 1 |
|
Nkx1-2
|
ENSMUSG00000048528.7 | NK1 homeobox 2 |
Activity of the Nkx1-1_Nkx1-2 motif across conditions
Conditions sorted by the z-value of the Nkx1-1_Nkx1-2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
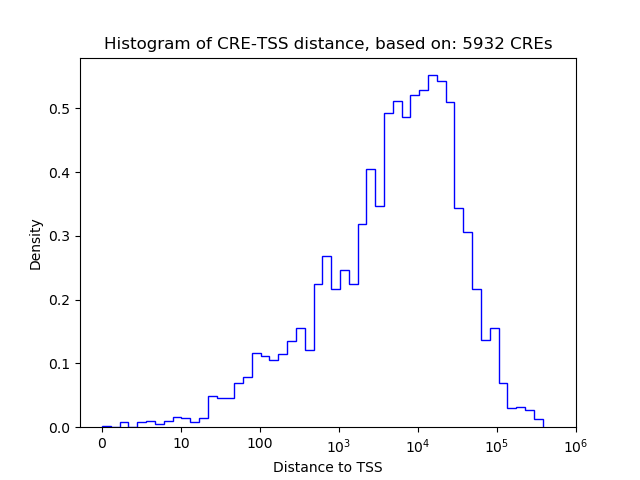
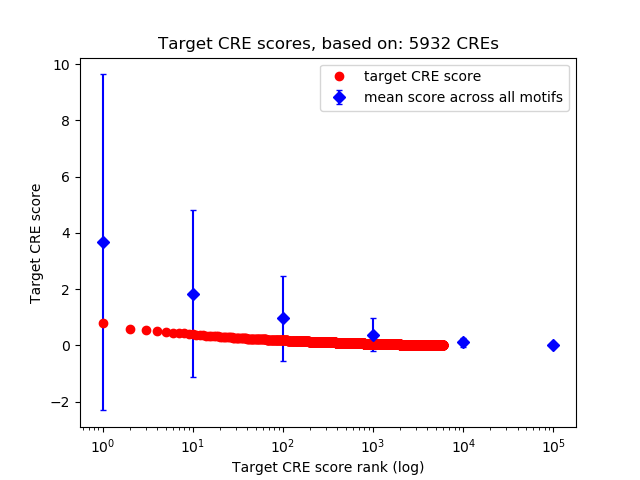
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_112530748_112531068 | 0.79 |
Gm27944 |
predicted gene, 27944 |
17461 |
0.15 |
| chr13_80895235_80895839 | 0.59 |
Arrdc3 |
arrestin domain containing 3 |
5019 |
0.18 |
| chr5_138080510_138080792 | 0.56 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr7_139559759_139559933 | 0.52 |
Nkx6-2 |
NK6 homeobox 2 |
22944 |
0.17 |
| chr3_105798169_105798320 | 0.49 |
Rap1a |
RAS-related protein 1a |
3041 |
0.15 |
| chr19_43971247_43971535 | 0.45 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr4_150598739_150598957 | 0.43 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
16567 |
0.18 |
| chr16_31998270_31998421 | 0.42 |
Senp5 |
SUMO/sentrin specific peptidase 5 |
4818 |
0.09 |
| chr7_26087500_26087684 | 0.41 |
Gm29920 |
predicted gene, 29920 |
12724 |
0.13 |
| chr11_30774536_30774692 | 0.40 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
2293 |
0.25 |
| chr7_39959522_39959690 | 0.38 |
Gm44992 |
predicted gene 44992 |
18468 |
0.21 |
| chr5_114553263_114553436 | 0.37 |
Gm13790 |
predicted gene 13790 |
2060 |
0.28 |
| chr13_58058115_58058280 | 0.36 |
Mir874 |
microRNA 874 |
34997 |
0.13 |
| chr13_60267432_60267612 | 0.35 |
Gm24999 |
predicted gene, 24999 |
26150 |
0.16 |
| chr3_97337526_97337699 | 0.33 |
Bcl9 |
B cell CLL/lymphoma 9 |
39695 |
0.14 |
| chr4_148040579_148040753 | 0.32 |
Mthfr |
methylenetetrahydrofolate reductase |
144 |
0.9 |
| chr11_108936283_108936686 | 0.32 |
Axin2 |
axin 2 |
2390 |
0.3 |
| chr17_54678232_54678387 | 0.31 |
Gm26291 |
predicted gene, 26291 |
26373 |
0.25 |
| chr1_89898377_89898561 | 0.31 |
Gbx2 |
gastrulation brain homeobox 2 |
30716 |
0.16 |
| chr1_24612181_24612351 | 0.31 |
Gm28439 |
predicted gene 28439 |
144 |
0.72 |
| chr2_75202592_75202875 | 0.31 |
Gm13653 |
predicted gene 13653 |
10456 |
0.15 |
| chr4_153224874_153225025 | 0.30 |
Gm13174 |
predicted gene 13174 |
6773 |
0.3 |
| chr6_50981986_50982138 | 0.30 |
Gm44402 |
predicted gene, 44402 |
14794 |
0.23 |
| chr3_10149901_10150052 | 0.30 |
Pmp2 |
peripheral myelin protein 2 |
33953 |
0.1 |
| chr6_28896411_28896583 | 0.29 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
16454 |
0.2 |
| chr3_28706532_28706683 | 0.29 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
8590 |
0.17 |
| chr6_121878156_121878361 | 0.28 |
Mug1 |
murinoglobulin 1 |
7319 |
0.2 |
| chr12_59160932_59161116 | 0.27 |
Mia2 |
MIA SH3 domain ER export factor 2 |
1866 |
0.28 |
| chr10_123843693_123843994 | 0.26 |
Gm18510 |
predicted gene, 18510 |
48055 |
0.2 |
| chr8_71699244_71699423 | 0.26 |
B3gnt3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
2456 |
0.13 |
| chr11_106013602_106013764 | 0.26 |
Kcnh6 |
potassium voltage-gated channel, subfamily H (eag-related), member 6 |
90 |
0.94 |
| chr6_97871999_97872401 | 0.26 |
Gm15531 |
predicted gene 15531 |
18339 |
0.23 |
| chr11_96551158_96551309 | 0.26 |
Skap1 |
src family associated phosphoprotein 1 |
61372 |
0.09 |
| chr9_112123334_112123526 | 0.25 |
Mir128-2 |
microRNA 128-2 |
4719 |
0.29 |
| chr9_83099037_83099215 | 0.25 |
Gm38398 |
predicted gene, 38398 |
902 |
0.5 |
| chr14_20177622_20177781 | 0.25 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
4108 |
0.16 |
| chr18_46722381_46722727 | 0.25 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
5475 |
0.14 |
| chr1_66862471_66862695 | 0.25 |
Acadl |
acyl-Coenzyme A dehydrogenase, long-chain |
694 |
0.5 |
| chr4_32507009_32507324 | 0.24 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
5661 |
0.25 |
| chr17_7284590_7284752 | 0.24 |
Rps6ka2 |
ribosomal protein S6 kinase, polypeptide 2 |
24035 |
0.14 |
| chr8_41052995_41053172 | 0.24 |
Gm16193 |
predicted gene 16193 |
27 |
0.96 |
| chr4_117190012_117190473 | 0.24 |
Gm25099 |
predicted gene, 25099 |
300 |
0.73 |
| chr6_112531173_112531324 | 0.23 |
Gm27944 |
predicted gene, 27944 |
17801 |
0.15 |
| chr1_21131249_21131442 | 0.23 |
Gm2693 |
predicted gene 2693 |
47643 |
0.1 |
| chr4_103934685_103934997 | 0.23 |
Gm12719 |
predicted gene 12719 |
4700 |
0.27 |
| chr2_29443856_29444159 | 0.23 |
Gm24976 |
predicted gene, 24976 |
15187 |
0.16 |
| chr2_119046481_119046794 | 0.23 |
Knl1 |
kinetochore scaffold 1 |
482 |
0.71 |
| chr11_111605518_111605669 | 0.23 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr11_20543948_20544116 | 0.23 |
Sertad2 |
SERTA domain containing 2 |
698 |
0.73 |
| chr2_146487304_146487455 | 0.23 |
Ralgapa2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
24520 |
0.22 |
| chr10_25595963_25596295 | 0.23 |
Gm36908 |
predicted gene, 36908 |
25434 |
0.15 |
| chr5_89416345_89416742 | 0.23 |
Gc |
vitamin D binding protein |
19085 |
0.24 |
| chr15_11380333_11380500 | 0.23 |
Tars |
threonyl-tRNA synthetase |
6929 |
0.25 |
| chr1_84186097_84186268 | 0.22 |
Gm18304 |
predicted gene, 18304 |
17994 |
0.2 |
| chr2_169724307_169724473 | 0.22 |
Tshz2 |
teashirt zinc finger family member 2 |
90714 |
0.08 |
| chr12_5668476_5668791 | 0.22 |
Gm48599 |
predicted gene, 48599 |
77101 |
0.1 |
| chr3_85551336_85551504 | 0.22 |
Gm42812 |
predicted gene 42812 |
1969 |
0.31 |
| chr14_21071028_21071191 | 0.22 |
Adk |
adenosine kinase |
5043 |
0.23 |
| chr3_85831893_85832058 | 0.21 |
Fam160a1 |
family with sequence similarity 160, member A1 |
14684 |
0.15 |
| chr11_23954708_23954885 | 0.21 |
Gm12062 |
predicted gene 12062 |
25942 |
0.16 |
| chr14_114090168_114090331 | 0.21 |
Gm18369 |
predicted gene, 18369 |
82194 |
0.11 |
| chr14_64865582_64865960 | 0.21 |
Gm37183 |
predicted gene, 37183 |
26512 |
0.14 |
| chr6_99210315_99210483 | 0.21 |
Foxp1 |
forkhead box P1 |
37937 |
0.21 |
| chr18_64346648_64346799 | 0.21 |
Onecut2 |
one cut domain, family member 2 |
6703 |
0.18 |
| chr3_144279874_144280028 | 0.21 |
Gm43446 |
predicted gene 43446 |
3860 |
0.25 |
| chr1_147365933_147366089 | 0.20 |
Gm9931 |
predicted gene 9931 |
84069 |
0.1 |
| chr5_145881335_145881631 | 0.20 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
1519 |
0.36 |
| chr2_34049474_34049625 | 0.20 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr1_36469911_36470078 | 0.20 |
Cnnm4 |
cyclin M4 |
1626 |
0.22 |
| chr12_83087732_83087912 | 0.20 |
Gm29530 |
predicted gene 29530 |
11052 |
0.17 |
| chr5_51511759_51511910 | 0.20 |
Gm43605 |
predicted gene 43605 |
2459 |
0.24 |
| chr8_77961865_77962061 | 0.20 |
Gm29895 |
predicted gene, 29895 |
82047 |
0.09 |
| chr5_135782110_135782261 | 0.20 |
Mdh2 |
malate dehydrogenase 2, NAD (mitochondrial) |
3463 |
0.13 |
| chr4_48152901_48153059 | 0.20 |
Stx17 |
syntaxin 17 |
28045 |
0.17 |
| chr7_14430420_14431121 | 0.20 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
5677 |
0.17 |
| chr8_125390183_125390334 | 0.20 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
102452 |
0.07 |
| chr16_31282098_31282249 | 0.20 |
Ppp1r2 |
protein phosphatase 1, regulatory inhibitor subunit 2 |
6896 |
0.11 |
| chr2_64915505_64915665 | 0.20 |
Grb14 |
growth factor receptor bound protein 14 |
566 |
0.84 |
| chr8_93959067_93959388 | 0.20 |
Gnao1 |
guanine nucleotide binding protein, alpha O |
3617 |
0.19 |
| chr9_30613779_30614132 | 0.20 |
Gm47716 |
predicted gene, 47716 |
18632 |
0.24 |
| chr11_95724615_95724802 | 0.20 |
Zfp652 |
zinc finger protein 652 |
11697 |
0.12 |
| chr10_7265928_7266272 | 0.20 |
Cnksr3 |
Cnksr family member 3 |
53863 |
0.15 |
| chr11_98339208_98339359 | 0.20 |
Ppp1r1b |
protein phosphatase 1, regulatory inhibitor subunit 1B |
9121 |
0.09 |
| chr11_100330372_100330542 | 0.19 |
Gast |
gastrin |
3950 |
0.09 |
| chr1_72226394_72226550 | 0.19 |
Gm25360 |
predicted gene, 25360 |
232 |
0.89 |
| chr15_77208076_77208227 | 0.19 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
4715 |
0.18 |
| chr12_102302828_102303218 | 0.19 |
Rin3 |
Ras and Rab interactor 3 |
19382 |
0.2 |
| chr13_98594899_98595134 | 0.19 |
Gm4815 |
predicted gene 4815 |
18485 |
0.12 |
| chr13_75722257_75722538 | 0.19 |
Gm48302 |
predicted gene, 48302 |
4882 |
0.15 |
| chr3_27431035_27431235 | 0.19 |
Ghsr |
growth hormone secretagogue receptor |
59784 |
0.12 |
| chr11_54355292_54355454 | 0.19 |
Gm12224 |
predicted gene 12224 |
1894 |
0.24 |
| chr15_89471642_89471822 | 0.19 |
Arsa |
arylsulfatase A |
2539 |
0.13 |
| chr7_84664728_84664917 | 0.19 |
Zfand6 |
zinc finger, AN1-type domain 6 |
14529 |
0.17 |
| chr7_122533976_122534175 | 0.18 |
Gm14389 |
predicted gene 14389 |
49655 |
0.11 |
| chr18_44633864_44634028 | 0.18 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
27719 |
0.19 |
| chr11_3983568_3983719 | 0.18 |
Gal3st1 |
galactose-3-O-sulfotransferase 1 |
7 |
0.95 |
| chr1_178325132_178325283 | 0.18 |
Hnrnpu |
heterogeneous nuclear ribonucleoprotein U |
1036 |
0.37 |
| chr11_63791140_63791492 | 0.18 |
Gm12288 |
predicted gene 12288 |
26984 |
0.21 |
| chr5_117125186_117125352 | 0.18 |
Taok3 |
TAO kinase 3 |
1032 |
0.43 |
| chr9_90136592_90136799 | 0.18 |
Tmem41b-ps |
transmembrane protein 41B, pseudogene |
17758 |
0.13 |
| chr1_140018825_140019186 | 0.18 |
Gm26048 |
predicted gene, 26048 |
15180 |
0.15 |
| chr6_112531381_112531532 | 0.18 |
Gm27944 |
predicted gene, 27944 |
18009 |
0.15 |
| chr10_51553900_51554061 | 0.18 |
Gm48787 |
predicted gene, 48787 |
2533 |
0.16 |
| chr7_140776294_140776445 | 0.18 |
Gm29799 |
predicted gene, 29799 |
8146 |
0.09 |
| chr4_63738674_63738850 | 0.18 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
6351 |
0.22 |
| chr15_83471603_83471756 | 0.17 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
7073 |
0.15 |
| chr2_14099979_14100130 | 0.17 |
Stam |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
25627 |
0.12 |
| chr18_10461260_10461439 | 0.17 |
Greb1l |
growth regulation by estrogen in breast cancer-like |
3124 |
0.25 |
| chr2_178711802_178711999 | 0.17 |
Cdh26 |
cadherin-like 26 |
251270 |
0.02 |
| chr2_167358690_167358858 | 0.17 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
9591 |
0.19 |
| chr5_76043835_76044206 | 0.17 |
Gm32780 |
predicted gene, 32780 |
922 |
0.54 |
| chr15_53316381_53316532 | 0.17 |
Ext1 |
exostosin glycosyltransferase 1 |
29203 |
0.23 |
| chr18_46553461_46553763 | 0.17 |
Ticam2 |
toll-like receptor adaptor molecule 2 |
20921 |
0.12 |
| chr8_48553802_48554548 | 0.17 |
Tenm3 |
teneurin transmembrane protein 3 |
1138 |
0.64 |
| chr12_33285610_33285770 | 0.17 |
Atxn7l1 |
ataxin 7-like 1 |
16825 |
0.2 |
| chr18_54868802_54868964 | 0.17 |
Gm50366 |
predicted gene, 50366 |
783 |
0.63 |
| chr6_121847052_121847216 | 0.17 |
Mug1 |
murinoglobulin 1 |
6030 |
0.23 |
| chr5_87124253_87124404 | 0.17 |
Gm42795 |
predicted gene 42795 |
9719 |
0.12 |
| chr19_57611799_57612005 | 0.17 |
Atrnl1 |
attractin like 1 |
868 |
0.62 |
| chr2_160865636_160865792 | 0.17 |
Zhx3 |
zinc fingers and homeoboxes 3 |
6083 |
0.14 |
| chr1_58203680_58204225 | 0.17 |
Aox4 |
aldehyde oxidase 4 |
6445 |
0.19 |
| chr17_5798546_5798723 | 0.17 |
3300005D01Rik |
RIKEN cDNA 3300005D01 gene |
23 |
0.97 |
| chr18_9811565_9811748 | 0.17 |
Gm50079 |
predicted gene, 50079 |
7128 |
0.15 |
| chr16_29988549_29988737 | 0.17 |
Gm1968 |
predicted gene 1968 |
9459 |
0.16 |
| chr4_105181181_105181508 | 0.17 |
Plpp3 |
phospholipid phosphatase 3 |
23997 |
0.24 |
| chr13_85126680_85126842 | 0.17 |
Gm4076 |
predicted gene 4076 |
753 |
0.65 |
| chr17_88614606_88614762 | 0.16 |
Gm9406 |
predicted gene 9406 |
6140 |
0.17 |
| chr16_55810073_55810224 | 0.16 |
Nfkbiz |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
8367 |
0.18 |
| chr17_49205213_49205366 | 0.16 |
Gm9586 |
predicted gene 9586 |
39531 |
0.16 |
| chr13_80896624_80897028 | 0.16 |
Arrdc3 |
arrestin domain containing 3 |
6308 |
0.17 |
| chr9_115401508_115401659 | 0.16 |
Gm9487 |
predicted gene 9487 |
3566 |
0.16 |
| chr19_38356216_38356572 | 0.16 |
Gm50150 |
predicted gene, 50150 |
13780 |
0.14 |
| chr16_24394028_24394581 | 0.16 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr11_53420114_53420567 | 0.16 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
2830 |
0.12 |
| chr5_67215059_67215221 | 0.16 |
Tmem33 |
transmembrane protein 33 |
45425 |
0.12 |
| chr16_76349002_76349359 | 0.16 |
Nrip1 |
nuclear receptor interacting protein 1 |
23857 |
0.18 |
| chr1_60862632_60862783 | 0.16 |
Gm38137 |
predicted gene, 38137 |
17631 |
0.12 |
| chr4_53104702_53105001 | 0.16 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
55044 |
0.11 |
| chr1_190928195_190928363 | 0.16 |
Angel2 |
angel homolog 2 |
213 |
0.92 |
| chrX_16816054_16816274 | 0.16 |
Maob |
monoamine oxidase B |
1111 |
0.65 |
| chrX_75129625_75129782 | 0.16 |
Mpp1 |
membrane protein, palmitoylated |
1141 |
0.32 |
| chr18_20654349_20654505 | 0.16 |
Gm16090 |
predicted gene 16090 |
10833 |
0.17 |
| chr12_17742392_17742683 | 0.16 |
Hpcal1 |
hippocalcin-like 1 |
14311 |
0.22 |
| chr9_105878487_105878719 | 0.16 |
Col6a5 |
collagen, type VI, alpha 5 |
3203 |
0.27 |
| chr13_112237981_112238162 | 0.15 |
Gm37427 |
predicted gene, 37427 |
18878 |
0.17 |
| chr7_127666140_127666314 | 0.15 |
Gm44760 |
predicted gene 44760 |
7993 |
0.08 |
| chr19_26600149_26600340 | 0.15 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
4806 |
0.21 |
| chr3_85821665_85821854 | 0.15 |
Fam160a1 |
family with sequence similarity 160, member A1 |
4468 |
0.2 |
| chr6_36281020_36281243 | 0.15 |
9330158H04Rik |
RIKEN cDNA 9330158H04 gene |
6775 |
0.29 |
| chr1_92180367_92180696 | 0.15 |
Hdac4 |
histone deacetylase 4 |
190 |
0.96 |
| chr10_82220492_82220673 | 0.15 |
Zfp938 |
zinc finger protein 938 |
20691 |
0.13 |
| chr3_121279413_121279568 | 0.15 |
Gm30517 |
predicted gene, 30517 |
1966 |
0.25 |
| chr18_37765813_37765974 | 0.15 |
Pcdhga12 |
protocadherin gamma subfamily A, 12 |
120 |
0.85 |
| chr14_28505648_28505933 | 0.15 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
110 |
0.97 |
| chr13_113535001_113535152 | 0.15 |
4921509O07Rik |
RIKEN cDNA 4921509O07 gene |
32232 |
0.12 |
| chr5_72404662_72404813 | 0.15 |
Gm19560 |
predicted gene, 19560 |
4707 |
0.18 |
| chr7_133052922_133053103 | 0.15 |
Ctbp2 |
C-terminal binding protein 2 |
22896 |
0.16 |
| chr10_75522174_75522329 | 0.15 |
Snrpd3 |
small nuclear ribonucleoprotein D3 |
4208 |
0.13 |
| chr1_43445213_43445518 | 0.15 |
Nck2 |
non-catalytic region of tyrosine kinase adaptor protein 2 |
386 |
0.87 |
| chr1_188853017_188853420 | 0.15 |
Gm25269 |
predicted gene, 25269 |
92742 |
0.08 |
| chr11_114375981_114376365 | 0.15 |
Gm11692 |
predicted gene 11692 |
19044 |
0.21 |
| chr4_134922832_134923008 | 0.15 |
Rsrp1 |
arginine/serine rich protein 1 |
672 |
0.64 |
| chr8_120230897_120231102 | 0.15 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
2543 |
0.23 |
| chr8_22510204_22510364 | 0.15 |
Slc20a2 |
solute carrier family 20, member 2 |
4058 |
0.18 |
| chr17_29094634_29094798 | 0.15 |
1700023B13Rik |
RIKEN cDNA 1700023B13 gene |
255 |
0.77 |
| chr1_187764201_187764362 | 0.15 |
AC121143.1 |
NADH dehydrogenase 2, mitochondrial (mt-Nd2) pseudogene |
51476 |
0.16 |
| chr12_84165735_84165922 | 0.15 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
5690 |
0.11 |
| chr2_163541251_163541470 | 0.15 |
Hnf4aos |
hepatic nuclear factor 4 alpha, opposite strand |
332 |
0.83 |
| chr12_75862976_75863134 | 0.14 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
10325 |
0.24 |
| chr11_95869540_95869834 | 0.14 |
Gm11534 |
predicted gene 11534 |
533 |
0.62 |
| chr5_93323705_93323875 | 0.14 |
Ccng2 |
cyclin G2 |
55082 |
0.1 |
| chr3_145621208_145621546 | 0.14 |
Znhit6 |
zinc finger, HIT type 6 |
25509 |
0.15 |
| chr1_4820222_4820413 | 0.14 |
Lypla1 |
lysophospholipase 1 |
12080 |
0.11 |
| chr5_63897626_63897962 | 0.14 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
20318 |
0.15 |
| chr5_65599782_65600155 | 0.14 |
Ube2k |
ubiquitin-conjugating enzyme E2K |
8372 |
0.1 |
| chr10_116221046_116221335 | 0.14 |
Ptprr |
protein tyrosine phosphatase, receptor type, R |
24794 |
0.18 |
| chr7_25916218_25916383 | 0.14 |
Cyp2b10 |
cytochrome P450, family 2, subfamily b, polypeptide 10 |
466 |
0.72 |
| chr16_20371861_20372035 | 0.14 |
Abcc5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
4694 |
0.18 |
| chr18_46712849_46713058 | 0.14 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
15076 |
0.12 |
| chr1_165760792_165760967 | 0.14 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
2867 |
0.13 |
| chr6_121890040_121890777 | 0.14 |
Mug1 |
murinoglobulin 1 |
4831 |
0.21 |
| chr4_81841873_81842024 | 0.14 |
n-R5s187 |
nuclear encoded rRNA 5S 187 |
76565 |
0.1 |
| chr2_69911096_69911271 | 0.14 |
Ubr3 |
ubiquitin protein ligase E3 component n-recognin 3 |
13880 |
0.13 |
| chr11_43217118_43217287 | 0.14 |
Gm12144 |
predicted gene 12144 |
15062 |
0.19 |
| chr6_50986659_50986810 | 0.14 |
Gm44402 |
predicted gene, 44402 |
19466 |
0.22 |
| chr10_115298774_115299175 | 0.14 |
Rab21 |
RAB21, member RAS oncogene family |
16617 |
0.14 |
| chr3_135842259_135842432 | 0.14 |
4933401H06Rik |
RIKEN cDNA 4933401H06 gene |
2076 |
0.25 |
| chr12_78215748_78215899 | 0.14 |
Gphn |
gephyrin |
10556 |
0.14 |
| chr7_123124873_123125354 | 0.14 |
Tnrc6a |
trinucleotide repeat containing 6a |
857 |
0.66 |
| chr3_86560323_86560494 | 0.14 |
Mab21l2 |
mab-21-like 2 |
11779 |
0.21 |
| chr13_37665370_37665760 | 0.14 |
AI463229 |
expressed sequence AI463229 |
977 |
0.42 |
| chr8_119912901_119913087 | 0.14 |
Usp10 |
ubiquitin specific peptidase 10 |
2137 |
0.24 |
| chr2_118318601_118318758 | 0.14 |
1700054M17Rik |
RIKEN cDNA 1700054M17 gene |
13765 |
0.13 |
| chr5_112908382_112908548 | 0.14 |
Myo18b |
myosin XVIIIb |
12103 |
0.19 |
| chr11_72737626_72737777 | 0.14 |
Ankfy1 |
ankyrin repeat and FYVE domain containing 1 |
22215 |
0.16 |
| chr11_107467010_107467274 | 0.13 |
Pitpnc1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
3557 |
0.15 |
| chr9_121928936_121929087 | 0.13 |
Gm47115 |
predicted gene, 47115 |
5069 |
0.09 |
| chr15_75803070_75803233 | 0.13 |
Zc3h3 |
zinc finger CCCH type containing 3 |
9708 |
0.12 |
| chr10_63060758_63060924 | 0.13 |
Pbld1 |
phenazine biosynthesis-like protein domain containing 1 |
112 |
0.93 |
| chr13_46466566_46466717 | 0.13 |
Cap2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
35207 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 0.2 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0014894 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 0.1 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0034819 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |