Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
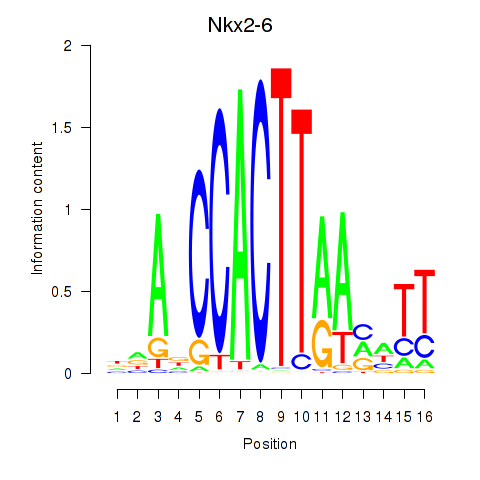
Results for Nkx2-6
Z-value: 1.04
Transcription factors associated with Nkx2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-6
|
ENSMUSG00000044186.9 | NK2 homeobox 6 |
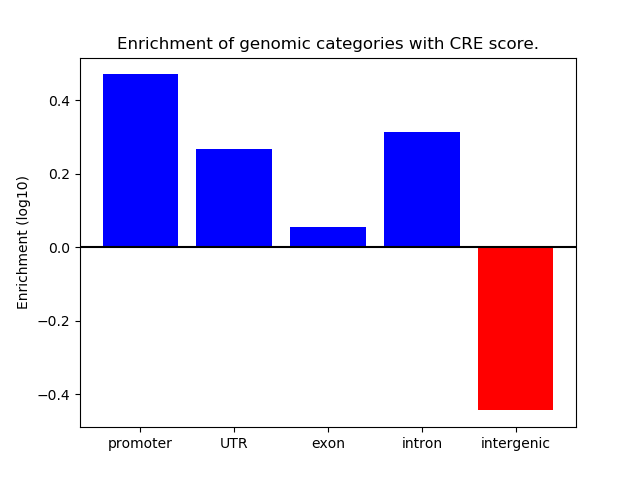
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_69171500_69171815 | Nkx2-6 | 145 | 0.567867 | -0.35 | 4.9e-01 | Click! |
Activity of the Nkx2-6 motif across conditions
Conditions sorted by the z-value of the Nkx2-6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
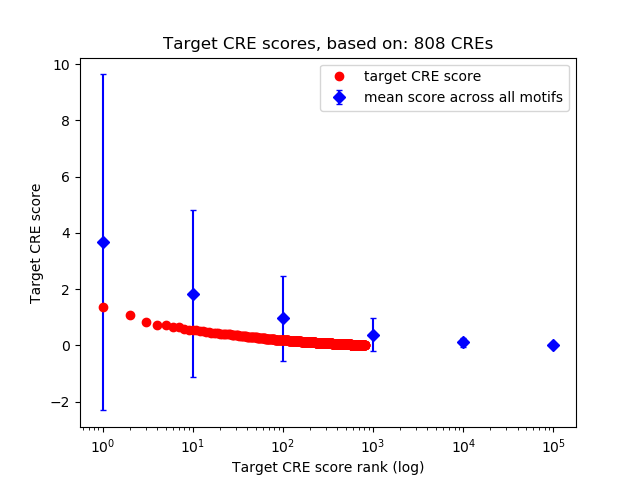
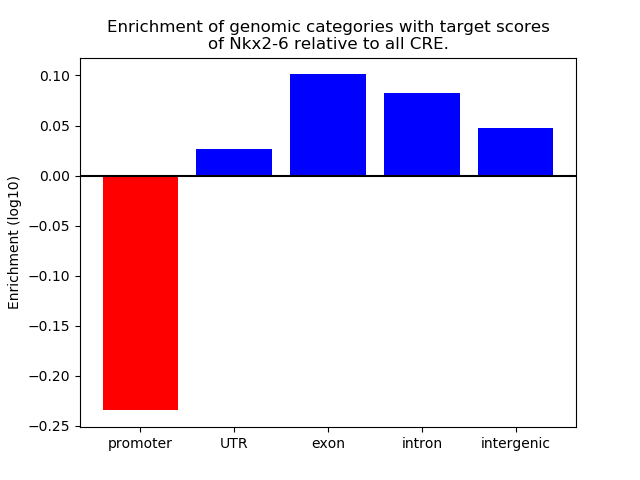
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_52319247_52319421 | 1.36 |
Neb |
nebulin |
15800 |
0.17 |
| chr13_41019879_41020036 | 1.07 |
Tmem14c |
transmembrane protein 14C |
3665 |
0.15 |
| chr7_126636776_126636927 | 0.84 |
Nupr1 |
nuclear protein transcription regulator 1 |
5990 |
0.08 |
| chr3_144617682_144617841 | 0.71 |
Rpl9-ps8 |
ribosomal protein L9, pseudogene 8 |
8203 |
0.15 |
| chr1_187214930_187215194 | 0.71 |
Spata17 |
spermatogenesis associated 17 |
359 |
0.55 |
| chr17_42900854_42901005 | 0.65 |
Cd2ap |
CD2-associated protein |
24264 |
0.24 |
| chr9_56260139_56260312 | 0.64 |
Peak1os |
pseudopodium-enriched atypical kinase 1, opposite strand |
2335 |
0.21 |
| chr11_16588986_16589143 | 0.59 |
Gm12663 |
predicted gene 12663 |
47002 |
0.12 |
| chr11_32659387_32659538 | 0.56 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
16558 |
0.21 |
| chr5_32328252_32328418 | 0.56 |
Gm15615 |
predicted gene 15615 |
16216 |
0.14 |
| chr2_148236402_148236558 | 0.53 |
Gm24221 |
predicted gene, 24221 |
85330 |
0.08 |
| chr3_68508770_68508933 | 0.52 |
Schip1 |
schwannomin interacting protein 1 |
14643 |
0.22 |
| chr19_45958655_45958833 | 0.52 |
Armh3 |
armadillo-like helical domain containing 3 |
22315 |
0.13 |
| chr2_35496007_35496180 | 0.49 |
Gm13445 |
predicted gene 13445 |
4574 |
0.15 |
| chr16_72452183_72452361 | 0.47 |
Gm49670 |
predicted gene, 49670 |
79831 |
0.11 |
| chr2_91216142_91216316 | 0.44 |
Acp2 |
acid phosphatase 2, lysosomal |
13301 |
0.09 |
| chr15_3380372_3380645 | 0.43 |
Ccdc152 |
coiled-coil domain containing 152 |
76982 |
0.1 |
| chr15_75111208_75111372 | 0.43 |
Ly6c2 |
lymphocyte antigen 6 complex, locus C2 |
407 |
0.75 |
| chr9_25525770_25525922 | 0.42 |
Gm25861 |
predicted gene, 25861 |
13359 |
0.19 |
| chr8_114151087_114151399 | 0.41 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
17601 |
0.25 |
| chr14_26156193_26156349 | 0.41 |
Gm2274 |
predicted gene 2274 |
235 |
0.89 |
| chr12_59040509_59040699 | 0.40 |
Gm22973 |
predicted gene, 22973 |
269 |
0.85 |
| chr11_90224957_90225225 | 0.40 |
Mmd |
monocyte to macrophage differentiation-associated |
24365 |
0.15 |
| chr7_68280697_68280888 | 0.39 |
Gm16157 |
predicted gene 16157 |
4198 |
0.16 |
| chr14_25876809_25876970 | 0.39 |
Anxa11os |
annexin A11, opposite strand |
6623 |
0.13 |
| chr14_26016579_26016740 | 0.39 |
Gm2260 |
predicted gene 2260 |
228 |
0.89 |
| chr6_121146935_121147121 | 0.38 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
16029 |
0.13 |
| chr5_129974617_129974800 | 0.37 |
Vkorc1l1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
2286 |
0.18 |
| chr11_16874843_16874994 | 0.37 |
Egfr |
epidermal growth factor receptor |
3232 |
0.26 |
| chr15_59391938_59392317 | 0.36 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
17840 |
0.18 |
| chr12_71863911_71864079 | 0.35 |
Gm7985 |
predicted gene 7985 |
16704 |
0.2 |
| chr11_69554847_69555031 | 0.35 |
Efnb3 |
ephrin B3 |
5266 |
0.08 |
| chr7_123369817_123370115 | 0.34 |
Arhgap17 |
Rho GTPase activating protein 17 |
51 |
0.68 |
| chr13_113901345_113901524 | 0.34 |
Arl15 |
ADP-ribosylation factor-like 15 |
106812 |
0.06 |
| chr8_84698956_84699121 | 0.33 |
Trmt1 |
tRNA methyltransferase 1 |
33 |
0.95 |
| chr5_122602369_122602541 | 0.33 |
Ift81 |
intraflagellar transport 81 |
1038 |
0.42 |
| chr6_29658788_29659068 | 0.33 |
Tspan33 |
tetraspanin 33 |
35294 |
0.13 |
| chr9_74793041_74793363 | 0.32 |
Gm22315 |
predicted gene, 22315 |
11132 |
0.18 |
| chr8_77961865_77962061 | 0.31 |
Gm29895 |
predicted gene, 29895 |
82047 |
0.09 |
| chr15_93744270_93744427 | 0.31 |
Gm30339 |
predicted gene, 30339 |
9935 |
0.21 |
| chr2_20909493_20909665 | 0.31 |
Arhgap21 |
Rho GTPase activating protein 21 |
5115 |
0.23 |
| chr11_22880864_22881030 | 0.31 |
Gm24917 |
predicted gene, 24917 |
12046 |
0.12 |
| chr15_62712455_62712777 | 0.31 |
Gm24810 |
predicted gene, 24810 |
59612 |
0.14 |
| chr16_22398929_22399080 | 0.30 |
Etv5 |
ets variant 5 |
2825 |
0.28 |
| chr17_28982482_28982633 | 0.30 |
Stk38 |
serine/threonine kinase 38 |
371 |
0.72 |
| chr6_53995717_53996030 | 0.30 |
4921529L05Rik |
RIKEN cDNA 4921529L05 gene |
13839 |
0.18 |
| chr3_156794209_156794402 | 0.29 |
Gm15578 |
predicted gene 15578 |
21732 |
0.19 |
| chr8_53780941_53781092 | 0.29 |
Gm19921 |
predicted gene, 19921 |
130723 |
0.05 |
| chr7_14430188_14430339 | 0.29 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
6184 |
0.17 |
| chr13_45960108_45960603 | 0.29 |
Atxn1 |
ataxin 1 |
4602 |
0.21 |
| chr13_17717656_17717837 | 0.28 |
Gm48621 |
predicted gene, 48621 |
7882 |
0.12 |
| chr13_28738845_28739011 | 0.28 |
Mir6368 |
microRNA 6368 |
28055 |
0.21 |
| chr3_18131799_18132081 | 0.27 |
Gm23686 |
predicted gene, 23686 |
45685 |
0.14 |
| chr6_92468512_92468696 | 0.27 |
Prickle2 |
prickle planar cell polarity protein 2 |
12788 |
0.26 |
| chr2_69137808_69137978 | 0.27 |
Nostrin |
nitric oxide synthase trafficker |
2093 |
0.33 |
| chr8_48730833_48730999 | 0.27 |
Tenm3 |
teneurin transmembrane protein 3 |
56226 |
0.15 |
| chr5_139871644_139871802 | 0.27 |
Gm42423 |
predicted gene 42423 |
24086 |
0.12 |
| chr1_85592768_85593352 | 0.26 |
Sp110 |
Sp110 nuclear body protein |
3869 |
0.12 |
| chr8_56313612_56313880 | 0.26 |
Gm45540 |
predicted gene 45540 |
2442 |
0.34 |
| chr10_84063624_84063775 | 0.25 |
Gm37908 |
predicted gene, 37908 |
1941 |
0.33 |
| chr5_33725824_33725998 | 0.25 |
Fgfr3 |
fibroblast growth factor receptor 3 |
2139 |
0.17 |
| chr6_31115031_31115198 | 0.25 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
4194 |
0.14 |
| chr10_25432440_25432591 | 0.25 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
84 |
0.97 |
| chr15_31357037_31357188 | 0.24 |
Ankrd33b |
ankyrin repeat domain 33B |
10420 |
0.16 |
| chr3_8924868_8925019 | 0.24 |
Mrps28 |
mitochondrial ribosomal protein S28 |
1025 |
0.53 |
| chr1_23484714_23484885 | 0.23 |
Gm7784 |
predicted gene 7784 |
5260 |
0.27 |
| chr19_21919071_21919233 | 0.23 |
Ldhb-ps |
lactate dehydrogenase B, pseudogene |
18228 |
0.18 |
| chr12_105034220_105034389 | 0.23 |
Glrx5 |
glutaredoxin 5 |
912 |
0.35 |
| chr5_144248287_144248466 | 0.23 |
2900089D17Rik |
RIKEN cDNA 2900089D17 gene |
1870 |
0.23 |
| chr6_28554965_28555164 | 0.23 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
6753 |
0.17 |
| chr13_68578198_68578368 | 0.23 |
Mtrr |
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
3611 |
0.19 |
| chr19_9027860_9028087 | 0.23 |
Ahnak |
AHNAK nucleoprotein (desmoyokin) |
28313 |
0.09 |
| chr1_168502271_168502436 | 0.23 |
Mir6348 |
microRNA 6348 |
11710 |
0.27 |
| chr3_69778568_69778734 | 0.22 |
Nmd3 |
NMD3 ribosome export adaptor |
32219 |
0.16 |
| chr13_110882966_110883130 | 0.22 |
Gm38397 |
predicted gene, 38397 |
20451 |
0.14 |
| chr8_33981301_33981581 | 0.22 |
Gm39157 |
predicted gene, 39157 |
4473 |
0.18 |
| chr10_111676325_111676608 | 0.22 |
Gm47864 |
predicted gene, 47864 |
26654 |
0.15 |
| chr10_68137101_68137496 | 0.22 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
672 |
0.78 |
| chr1_60824712_60825126 | 0.21 |
Gm11581 |
predicted gene 11581 |
15803 |
0.12 |
| chr3_98056406_98056789 | 0.21 |
Gm42819 |
predicted gene 42819 |
25910 |
0.16 |
| chr5_110802281_110802488 | 0.20 |
Ulk1 |
unc-51 like kinase 1 |
6550 |
0.11 |
| chr16_87690191_87690394 | 0.20 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
8653 |
0.22 |
| chr16_52322703_52322854 | 0.20 |
Alcam |
activated leukocyte cell adhesion molecule |
25854 |
0.26 |
| chr9_40773794_40774087 | 0.20 |
Clmp |
CXADR-like membrane protein |
154 |
0.91 |
| chr3_79587078_79587240 | 0.20 |
Ppid |
peptidylprolyl isomerase D (cyclophilin D) |
4183 |
0.13 |
| chr16_11638892_11639072 | 0.20 |
Gm4279 |
predicted gene 4279 |
52875 |
0.15 |
| chr4_49536222_49536402 | 0.20 |
Aldob |
aldolase B, fructose-bisphosphate |
2554 |
0.2 |
| chr16_30664084_30664254 | 0.20 |
Fam43a |
family with sequence similarity 43, member A |
64446 |
0.1 |
| chr1_133493056_133493228 | 0.20 |
Gm8596 |
predicted gene 8596 |
36337 |
0.14 |
| chr7_119668193_119668569 | 0.20 |
Acsm4 |
acyl-CoA synthetase medium-chain family member 4 |
21645 |
0.1 |
| chr17_72933196_72933347 | 0.20 |
Lbh |
limb-bud and heart |
12083 |
0.23 |
| chr8_36692402_36692571 | 0.19 |
Dlc1 |
deleted in liver cancer 1 |
40568 |
0.2 |
| chr6_87552153_87552313 | 0.19 |
Gm44198 |
predicted gene, 44198 |
18826 |
0.13 |
| chr17_53565206_53565384 | 0.19 |
Kat2b |
K(lysine) acetyltransferase 2B |
1566 |
0.33 |
| chr10_99384294_99384806 | 0.19 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
18240 |
0.17 |
| chr4_40156775_40156949 | 0.19 |
Aco1 |
aconitase 1 |
6857 |
0.2 |
| chr17_69396159_69396341 | 0.19 |
Gm49894 |
predicted gene, 49894 |
12042 |
0.14 |
| chr9_21692583_21692969 | 0.19 |
Gm26511 |
predicted gene, 26511 |
30674 |
0.1 |
| chr10_128231734_128232048 | 0.19 |
Timeless |
timeless circadian clock 1 |
174 |
0.87 |
| chr7_19668878_19669149 | 0.18 |
Clptm1 |
cleft lip and palate associated transmembrane protein 1 |
3980 |
0.09 |
| chr2_79977386_79977747 | 0.18 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
49775 |
0.17 |
| chr13_52037190_52037541 | 0.18 |
Gm37872 |
predicted gene, 37872 |
15438 |
0.21 |
| chr5_137054912_137055067 | 0.18 |
Ap1s1 |
adaptor protein complex AP-1, sigma 1 |
8854 |
0.1 |
| chr3_67677368_67677587 | 0.18 |
Gm37601 |
predicted gene, 37601 |
7275 |
0.2 |
| chr16_91765796_91766068 | 0.18 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
9833 |
0.16 |
| chr15_69043517_69043680 | 0.18 |
Gm49422 |
predicted gene, 49422 |
84067 |
0.09 |
| chr7_73430637_73430788 | 0.18 |
Gm44645 |
predicted gene 44645 |
8462 |
0.12 |
| chr10_80050367_80050530 | 0.18 |
Gpx4 |
glutathione peroxidase 4 |
3040 |
0.12 |
| chr19_55276564_55276746 | 0.18 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
3785 |
0.21 |
| chr11_75750659_75750835 | 0.17 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
857 |
0.56 |
| chr6_37473792_37473962 | 0.17 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
31731 |
0.19 |
| chr5_148740765_148741271 | 0.17 |
2210417A02Rik |
RIKEN cDNA 2210417A02 gene |
822 |
0.56 |
| chr1_62730735_62730912 | 0.17 |
Gm29083 |
predicted gene 29083 |
12750 |
0.18 |
| chr3_109255070_109255263 | 0.17 |
Gm13865 |
predicted gene 13865 |
62299 |
0.11 |
| chr4_62696097_62696258 | 0.17 |
Rgs3 |
regulator of G-protein signaling 3 |
3289 |
0.21 |
| chr4_136501510_136501674 | 0.17 |
Luzp1 |
leucine zipper protein 1 |
19803 |
0.14 |
| chr14_54653654_54653818 | 0.17 |
Acin1 |
apoptotic chromatin condensation inducer 1 |
35 |
0.93 |
| chr11_51753632_51753825 | 0.17 |
Sec24a |
Sec24 related gene family, member A (S. cerevisiae) |
2483 |
0.22 |
| chr1_138992310_138992466 | 0.17 |
Gm16305 |
predicted gene 16305 |
319 |
0.81 |
| chr14_117130450_117130601 | 0.17 |
Gpc6 |
glypican 6 |
204589 |
0.03 |
| chr5_119675143_119675355 | 0.17 |
Tbx3 |
T-box 3 |
196 |
0.92 |
| chr5_51552225_51552395 | 0.17 |
Ppargc1a |
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
1561 |
0.41 |
| chr14_62878202_62878375 | 0.17 |
n-R5s47 |
nuclear encoded rRNA 5S 47 |
8574 |
0.14 |
| chr6_21827909_21828060 | 0.17 |
Tspan12 |
tetraspanin 12 |
23844 |
0.17 |
| chr4_128840401_128840595 | 0.16 |
Gm12968 |
predicted gene 12968 |
5699 |
0.17 |
| chr15_57913180_57913355 | 0.16 |
Tbc1d31 |
TBC1 domain family, member 31 |
262 |
0.92 |
| chr1_170640374_170640534 | 0.16 |
Olfml2b |
olfactomedin-like 2B |
4078 |
0.21 |
| chr3_97076104_97076274 | 0.16 |
4930573H18Rik |
RIKEN cDNA 4930573H18 gene |
16596 |
0.15 |
| chr10_20005611_20005762 | 0.16 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
54631 |
0.13 |
| chr15_9596764_9597341 | 0.16 |
Il7r |
interleukin 7 receptor |
66876 |
0.12 |
| chr15_75048041_75048215 | 0.16 |
Ly6c1 |
lymphocyte antigen 6 complex, locus C1 |
541 |
0.61 |
| chr18_16753631_16753792 | 0.16 |
Gm15485 |
predicted gene 15485 |
24978 |
0.2 |
| chr1_77213552_77213762 | 0.15 |
Gm38265 |
predicted gene, 38265 |
66665 |
0.12 |
| chr14_63150536_63150699 | 0.15 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
14308 |
0.13 |
| chr3_7648648_7649046 | 0.15 |
Il7 |
interleukin 7 |
35087 |
0.17 |
| chr15_58977395_58977546 | 0.15 |
Mtss1 |
MTSS I-BAR domain containing 1 |
4931 |
0.17 |
| chr7_80741087_80741271 | 0.15 |
Iqgap1 |
IQ motif containing GTPase activating protein 1 |
2701 |
0.24 |
| chr4_139631247_139631408 | 0.15 |
Aldh4a1 |
aldehyde dehydrogenase 4 family, member A1 |
8218 |
0.14 |
| chr2_53063355_53063523 | 0.15 |
Prpf40a |
pre-mRNA processing factor 40A |
81870 |
0.1 |
| chr7_65338015_65338188 | 0.15 |
Tjp1 |
tight junction protein 1 |
4696 |
0.24 |
| chr4_97752078_97752249 | 0.15 |
Gm12676 |
predicted gene 12676 |
5226 |
0.23 |
| chr9_97490330_97490709 | 0.15 |
Gm25607 |
predicted gene, 25607 |
50892 |
0.16 |
| chr7_16583531_16584108 | 0.15 |
Gm29443 |
predicted gene 29443 |
30005 |
0.08 |
| chr8_61132865_61133032 | 0.15 |
Gm16179 |
predicted gene 16179 |
20446 |
0.16 |
| chr3_89113148_89113310 | 0.15 |
Gm29704 |
predicted gene, 29704 |
3495 |
0.09 |
| chr6_58832580_58832969 | 0.15 |
Herc3 |
hect domain and RLD 3 |
913 |
0.62 |
| chr1_107942021_107942201 | 0.15 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
5402 |
0.2 |
| chr12_73715979_73716146 | 0.15 |
Prkch |
protein kinase C, eta |
67141 |
0.1 |
| chr6_99656184_99656335 | 0.14 |
Gm44104 |
predicted gene, 44104 |
5959 |
0.16 |
| chr6_124509130_124509281 | 0.14 |
C1ra |
complement component 1, r subcomponent A |
3200 |
0.12 |
| chr5_115530819_115531136 | 0.14 |
Pxn |
paxillin |
10672 |
0.09 |
| chr8_46540344_46540495 | 0.14 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
9355 |
0.15 |
| chr18_36661287_36661681 | 0.14 |
Eif4ebp3 |
eukaryotic translation initiation factor 4E binding protein 3 |
2576 |
0.13 |
| chr10_83637415_83637787 | 0.14 |
Appl2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
158 |
0.96 |
| chr9_119309190_119309381 | 0.14 |
Gm22729 |
predicted gene, 22729 |
2528 |
0.19 |
| chr17_86911957_86912124 | 0.14 |
Tmem247 |
transmembrane protein 247 |
5308 |
0.16 |
| chr5_113133182_113133354 | 0.14 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
4718 |
0.12 |
| chr17_43105938_43106161 | 0.14 |
E130008D07Rik |
RIKEN cDNA E130008D07 gene |
52147 |
0.16 |
| chr9_120048495_120048762 | 0.13 |
Cx3cr1 |
chemokine (C-X3-C motif) receptor 1 |
19655 |
0.08 |
| chr19_8966037_8966203 | 0.13 |
Eef1g |
eukaryotic translation elongation factor 1 gamma |
921 |
0.31 |
| chr6_72416544_72416700 | 0.13 |
Ggcx |
gamma-glutamyl carboxylase |
1233 |
0.31 |
| chr5_133173380_133173603 | 0.13 |
Gm42625 |
predicted gene 42625 |
269851 |
0.02 |
| chr19_10268064_10268219 | 0.13 |
Dagla |
diacylglycerol lipase, alpha |
11270 |
0.13 |
| chr16_44063056_44063226 | 0.13 |
Gramd1c |
GRAM domain containing 1C |
184 |
0.95 |
| chr5_117979355_117979506 | 0.13 |
Fbxo21 |
F-box protein 21 |
106 |
0.95 |
| chr13_50051310_50051474 | 0.13 |
Gm48052 |
predicted gene, 48052 |
387 |
0.79 |
| chr5_99388868_99389019 | 0.13 |
Gm35394 |
predicted gene, 35394 |
114848 |
0.05 |
| chr11_98874019_98874180 | 0.13 |
Gm23640 |
predicted gene, 23640 |
3600 |
0.13 |
| chr15_96708796_96708976 | 0.13 |
Gm38144 |
predicted gene, 38144 |
8713 |
0.18 |
| chr5_77100130_77100295 | 0.13 |
Hopx |
HOP homeobox |
4935 |
0.14 |
| chr13_16715662_16715832 | 0.13 |
Gm7537 |
predicted gene 7537 |
76257 |
0.11 |
| chr12_72047672_72047836 | 0.13 |
Gm7986 |
predicted gene 7986 |
1505 |
0.36 |
| chr7_126109278_126109457 | 0.13 |
Xpo6 |
exportin 6 |
684 |
0.65 |
| chr1_133921507_133921777 | 0.13 |
Prelp |
proline arginine-rich end leucine-rich repeat |
228 |
0.9 |
| chr16_35309712_35309912 | 0.13 |
Gm49706 |
predicted gene, 49706 |
13254 |
0.15 |
| chr3_89770512_89770671 | 0.12 |
4632404H12Rik |
RIKEN cDNA 4632404H12 gene |
2200 |
0.18 |
| chr11_35007192_35007387 | 0.12 |
Gm25799 |
predicted gene, 25799 |
108272 |
0.06 |
| chr6_100185590_100185876 | 0.12 |
2010109P13Rik |
RIKEN cDNA 2010109P13 gene |
41919 |
0.13 |
| chr14_27338148_27338304 | 0.12 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
2160 |
0.35 |
| chr19_44064101_44064296 | 0.12 |
Erlin1 |
ER lipid raft associated 1 |
3485 |
0.17 |
| chr10_8830122_8830414 | 0.12 |
Gm25410 |
predicted gene, 25410 |
6121 |
0.2 |
| chr5_140644827_140644989 | 0.12 |
Ttyh3 |
tweety family member 3 |
4089 |
0.17 |
| chr17_65784660_65784842 | 0.12 |
Ppp4r1 |
protein phosphatase 4, regulatory subunit 1 |
1152 |
0.47 |
| chr11_84199091_84199285 | 0.12 |
Acaca |
acetyl-Coenzyme A carboxylase alpha |
3750 |
0.22 |
| chr1_48351528_48351702 | 0.12 |
Gm5974 |
predicted gene 5974 |
36764 |
0.22 |
| chrX_11130477_11130658 | 0.12 |
Gm14481 |
predicted gene 14481 |
90040 |
0.07 |
| chr7_80445419_80445914 | 0.12 |
Blm |
Bloom syndrome, RecQ like helicase |
11706 |
0.12 |
| chr19_6390399_6390550 | 0.12 |
Pygm |
muscle glycogen phosphorylase |
2237 |
0.14 |
| chr5_144904863_144905014 | 0.12 |
Smurf1 |
SMAD specific E3 ubiquitin protein ligase 1 |
6944 |
0.16 |
| chr16_25011043_25011318 | 0.11 |
A230028O05Rik |
RIKEN cDNA A230028O05 gene |
48459 |
0.18 |
| chr1_164703629_164703781 | 0.11 |
Gm37853 |
predicted gene, 37853 |
5472 |
0.17 |
| chr5_34408526_34408716 | 0.11 |
Fam193a |
family with sequence homology 193, member A |
17716 |
0.13 |
| chr1_161253186_161253391 | 0.11 |
Prdx6 |
peroxiredoxin 6 |
2069 |
0.29 |
| chr6_119352807_119352958 | 0.11 |
Cacna2d4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
20320 |
0.17 |
| chr8_56292788_56292973 | 0.11 |
Hpgd |
hydroxyprostaglandin dehydrogenase 15 (NAD) |
1705 |
0.43 |
| chr1_39372307_39372514 | 0.11 |
Tbc1d8 |
TBC1 domain family, member 8 |
23 |
0.97 |
| chr13_81328551_81328735 | 0.11 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
14193 |
0.26 |
| chr17_29579809_29579984 | 0.11 |
Tbc1d22b |
TBC1 domain family, member 22B |
7473 |
0.09 |
| chr4_100914770_100914929 | 0.11 |
Cachd1 |
cache domain containing 1 |
79762 |
0.09 |
| chr6_92219535_92219693 | 0.11 |
Rbsn |
rabenosyn, RAB effector |
4689 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |