Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
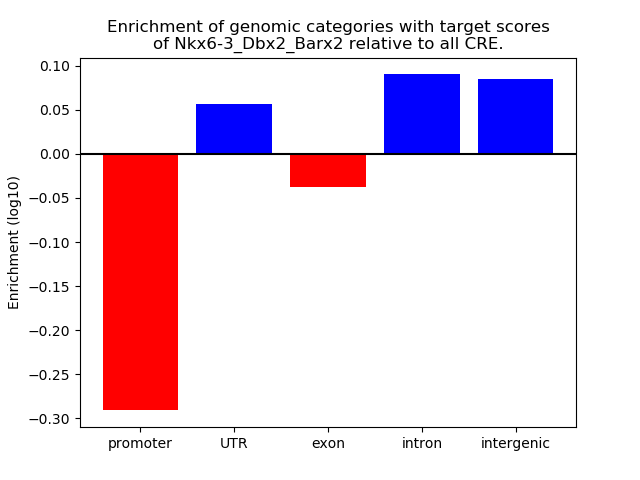
Results for Nkx6-3_Dbx2_Barx2
Z-value: 1.10
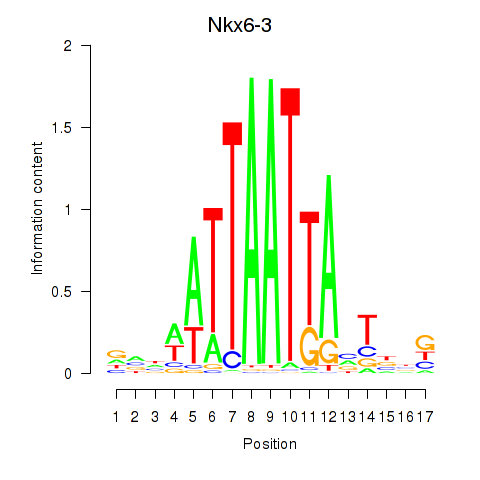
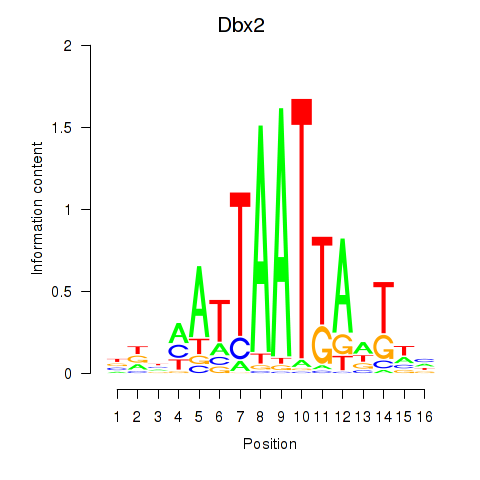
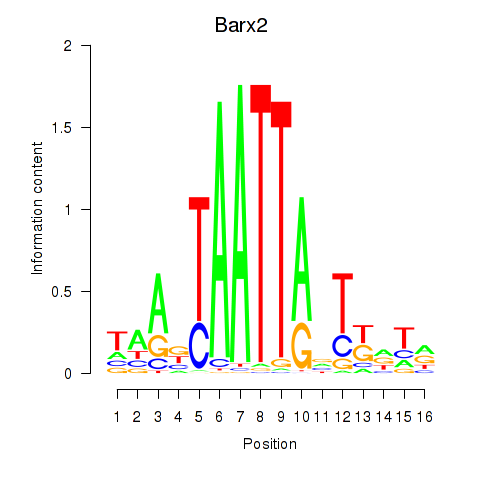
Transcription factors associated with Nkx6-3_Dbx2_Barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-3
|
ENSMUSG00000063672.6 | NK6 homeobox 3 |
|
Dbx2
|
ENSMUSG00000045608.6 | developing brain homeobox 2 |
|
Barx2
|
ENSMUSG00000032033.10 | BarH-like homeobox 2 |
Activity of the Nkx6-3_Dbx2_Barx2 motif across conditions
Conditions sorted by the z-value of the Nkx6-3_Dbx2_Barx2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
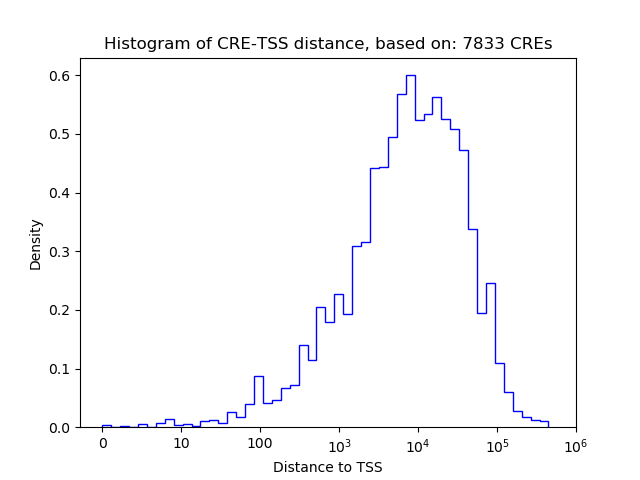
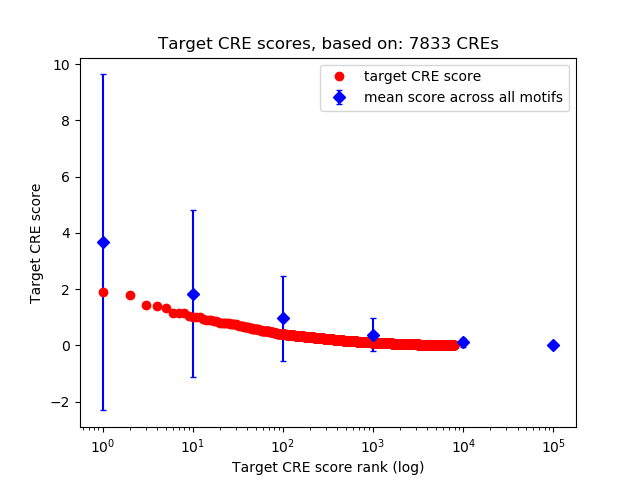
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_134708873_134709037 | 1.91 |
Gm14037 |
predicted gene 14037 |
22152 |
0.2 |
| chr11_16883025_16883203 | 1.80 |
Egfr |
epidermal growth factor receptor |
4964 |
0.23 |
| chr2_107628721_107628872 | 1.43 |
Gm9864 |
predicted gene 9864 |
28257 |
0.26 |
| chr6_144895356_144895507 | 1.39 |
Gm22792 |
predicted gene, 22792 |
91799 |
0.07 |
| chr1_100298746_100298897 | 1.31 |
Gm29667 |
predicted gene 29667 |
16030 |
0.18 |
| chr19_26823641_26823967 | 1.17 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
103 |
0.97 |
| chr8_25609907_25610083 | 1.16 |
Gm23184 |
predicted gene, 23184 |
563 |
0.58 |
| chr11_16798476_16798627 | 1.16 |
Egfros |
epidermal growth factor receptor, opposite strand |
32151 |
0.16 |
| chr3_148832133_148832284 | 1.03 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
1720 |
0.51 |
| chr2_73628966_73629117 | 1.02 |
Chn1 |
chimerin 1 |
3301 |
0.21 |
| chr13_63665698_63665976 | 1.01 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr7_140722780_140722931 | 1.01 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
1573 |
0.22 |
| chr16_87627804_87628113 | 0.93 |
Gm22808 |
predicted gene, 22808 |
6838 |
0.21 |
| chr14_86300654_86300845 | 0.92 |
Gm32815 |
predicted gene, 32815 |
1015 |
0.44 |
| chr12_104083020_104083260 | 0.90 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr4_44777019_44777170 | 0.90 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
16650 |
0.16 |
| chr12_51274477_51274668 | 0.88 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
22915 |
0.22 |
| chr10_24362567_24362800 | 0.86 |
Gm15271 |
predicted gene 15271 |
84807 |
0.08 |
| chr19_44401904_44402310 | 0.83 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
4583 |
0.17 |
| chr9_116136676_116136901 | 0.81 |
Tgfbr2 |
transforming growth factor, beta receptor II |
38477 |
0.15 |
| chr2_24040580_24040761 | 0.80 |
Hnmt |
histamine N-methyltransferase |
8196 |
0.23 |
| chr4_123990670_123991070 | 0.79 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr13_118829008_118829159 | 0.79 |
Gm47335 |
predicted gene, 47335 |
29331 |
0.22 |
| chr4_65080374_65080537 | 0.79 |
Pappa |
pregnancy-associated plasma protein A |
43719 |
0.19 |
| chr11_16885540_16885691 | 0.78 |
Egfr |
epidermal growth factor receptor |
7465 |
0.21 |
| chr3_111344475_111344661 | 0.77 |
Gm9314 |
predicted gene 9314 |
21711 |
0.29 |
| chr9_122826708_122826859 | 0.77 |
Gm35549 |
predicted gene, 35549 |
17103 |
0.1 |
| chr7_107574446_107574681 | 0.77 |
Olfml1 |
olfactomedin-like 1 |
6797 |
0.16 |
| chr10_69217231_69217448 | 0.76 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2037 |
0.3 |
| chr3_60525375_60525543 | 0.76 |
Mbnl1 |
muscleblind like splicing factor 1 |
2669 |
0.31 |
| chr19_40153747_40153898 | 0.73 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr11_16758574_16758725 | 0.71 |
Egfr |
epidermal growth factor receptor |
6419 |
0.21 |
| chr17_71222209_71222384 | 0.70 |
Lpin2 |
lipin 2 |
9137 |
0.17 |
| chr9_74328635_74328791 | 0.68 |
Gm24141 |
predicted gene, 24141 |
33897 |
0.17 |
| chr13_28877494_28877645 | 0.68 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
5887 |
0.2 |
| chr2_42022610_42022815 | 0.68 |
Gm13461 |
predicted gene 13461 |
33883 |
0.23 |
| chr3_18250842_18251072 | 0.66 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
7619 |
0.24 |
| chr9_74872523_74872674 | 0.66 |
Onecut1 |
one cut domain, family member 1 |
6114 |
0.16 |
| chr12_57537784_57538085 | 0.66 |
Foxa1 |
forkhead box A1 |
8187 |
0.15 |
| chr9_74885383_74885534 | 0.66 |
Onecut1 |
one cut domain, family member 1 |
18974 |
0.14 |
| chr8_94394237_94394701 | 0.65 |
Herpud1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
938 |
0.39 |
| chr1_84993315_84993466 | 0.63 |
Gm29284 |
predicted gene 29284 |
358 |
0.79 |
| chr5_52945521_52945741 | 0.62 |
Gm23532 |
predicted gene, 23532 |
13831 |
0.15 |
| chr3_97649987_97650151 | 0.61 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
8124 |
0.13 |
| chr8_73953761_73954148 | 0.61 |
Gm7948 |
predicted gene 7948 |
82261 |
0.11 |
| chr2_134500653_134500804 | 0.59 |
Hao1 |
hydroxyacid oxidase 1, liver |
53579 |
0.17 |
| chr18_54739961_54740121 | 0.58 |
Gm5821 |
predicted gene 5821 |
26091 |
0.21 |
| chr2_36201411_36201590 | 0.58 |
Gm13429 |
predicted gene 13429 |
814 |
0.51 |
| chr3_61217484_61217722 | 0.57 |
Gm37719 |
predicted gene, 37719 |
73843 |
0.1 |
| chr2_35184678_35184843 | 0.57 |
Rab14 |
RAB14, member RAS oncogene family |
612 |
0.69 |
| chr11_16839622_16839885 | 0.57 |
Egfros |
epidermal growth factor receptor, opposite strand |
9051 |
0.21 |
| chr13_24360335_24360523 | 0.57 |
Gm11342 |
predicted gene 11342 |
15521 |
0.12 |
| chr4_123990393_123990580 | 0.56 |
Gm12902 |
predicted gene 12902 |
64252 |
0.08 |
| chr14_69628830_69628981 | 0.56 |
Gm27177 |
predicted gene 27177 |
11369 |
0.11 |
| chr1_151718954_151719119 | 0.54 |
2810414N06Rik |
RIKEN cDNA 2810414N06 gene |
35982 |
0.14 |
| chr16_42947764_42948084 | 0.54 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
7707 |
0.21 |
| chr13_91208892_91209073 | 0.53 |
Gm17450 |
predicted gene, 17450 |
11470 |
0.19 |
| chr4_109104769_109104920 | 0.53 |
Osbpl9 |
oxysterol binding protein-like 9 |
3187 |
0.27 |
| chr10_87932167_87932318 | 0.52 |
Tyms-ps |
thymidylate synthase, pseudogene |
34605 |
0.14 |
| chr16_11406443_11406626 | 0.51 |
Snx29 |
sorting nexin 29 |
123 |
0.97 |
| chr17_64607771_64607970 | 0.51 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr11_84198789_84199054 | 0.50 |
Acaca |
acetyl-Coenzyme A carboxylase alpha |
3483 |
0.22 |
| chr3_118443694_118443845 | 0.50 |
Gm9916 |
predicted gene 9916 |
8999 |
0.15 |
| chr7_110192854_110193204 | 0.50 |
Swap70 |
SWA-70 protein |
28682 |
0.13 |
| chr17_80022007_80022158 | 0.50 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr16_65665507_65665824 | 0.50 |
Gm49633 |
predicted gene, 49633 |
52246 |
0.14 |
| chr10_8642632_8642823 | 0.50 |
Gm24374 |
predicted gene, 24374 |
207 |
0.96 |
| chr4_45800810_45800979 | 0.50 |
Aldh1b1 |
aldehyde dehydrogenase 1 family, member B1 |
1773 |
0.28 |
| chr14_18369164_18369315 | 0.49 |
Ube2e1 |
ubiquitin-conjugating enzyme E2E 1 |
37380 |
0.15 |
| chr18_39816253_39816404 | 0.49 |
Pabpc2 |
poly(A) binding protein, cytoplasmic 2 |
42831 |
0.17 |
| chr4_121078016_121078199 | 0.49 |
Zmpste24 |
zinc metallopeptidase, STE24 |
369 |
0.74 |
| chr11_35015732_35015915 | 0.49 |
Slit3 |
slit guidance ligand 3 |
105401 |
0.07 |
| chr16_43169836_43170008 | 0.48 |
Gm15712 |
predicted gene 15712 |
14651 |
0.21 |
| chr2_28618402_28618756 | 0.48 |
Gfi1b |
growth factor independent 1B |
3174 |
0.15 |
| chr13_80892366_80892552 | 0.47 |
Arrdc3 |
arrestin domain containing 3 |
1941 |
0.29 |
| chr3_149234408_149234559 | 0.47 |
Gm10287 |
predicted gene 10287 |
8738 |
0.21 |
| chr3_133759501_133760150 | 0.46 |
Gm6135 |
prediticted gene 6135 |
31679 |
0.18 |
| chr12_110576051_110576208 | 0.46 |
B930059L03Rik |
RIKEN cDNA B930059L03 gene |
14383 |
0.14 |
| chr3_18167606_18167757 | 0.45 |
Gm23686 |
predicted gene, 23686 |
9944 |
0.24 |
| chr11_97383112_97383279 | 0.45 |
Socs7 |
suppressor of cytokine signaling 7 |
5758 |
0.15 |
| chr1_56979180_56979331 | 0.44 |
Satb2 |
special AT-rich sequence binding protein 2 |
605 |
0.68 |
| chr13_44437957_44438156 | 0.44 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
1656 |
0.27 |
| chr12_95589166_95589349 | 0.43 |
Gm22246 |
predicted gene, 22246 |
30561 |
0.21 |
| chr9_98418368_98418712 | 0.43 |
Rbp1 |
retinol binding protein 1, cellular |
4421 |
0.24 |
| chr3_95872352_95872534 | 0.42 |
C920021L13Rik |
RIKEN cDNA C920021L13 gene |
912 |
0.26 |
| chrX_85734642_85734808 | 0.42 |
Gk |
glycerol kinase |
2885 |
0.21 |
| chr7_67578755_67579151 | 0.42 |
Lrrc28 |
leucine rich repeat containing 28 |
797 |
0.57 |
| chr11_16882054_16882234 | 0.42 |
Egfr |
epidermal growth factor receptor |
3994 |
0.24 |
| chr18_39438249_39438499 | 0.42 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
48858 |
0.13 |
| chr6_108253104_108253265 | 0.41 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
226 |
0.96 |
| chr12_69759378_69759543 | 0.41 |
Mir681 |
microRNA 681 |
4484 |
0.15 |
| chr6_16797841_16798030 | 0.41 |
Gm36669 |
predicted gene, 36669 |
20411 |
0.22 |
| chr10_86350127_86350278 | 0.41 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
47348 |
0.13 |
| chr2_13522226_13522421 | 0.41 |
Trdmt1 |
tRNA aspartic acid methyltransferase 1 |
22327 |
0.19 |
| chr12_80336463_80336614 | 0.41 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
40154 |
0.1 |
| chr13_16574847_16575001 | 0.41 |
Gm48497 |
predicted gene, 48497 |
41103 |
0.17 |
| chr2_122251200_122251358 | 0.41 |
Sord |
sorbitol dehydrogenase |
16530 |
0.1 |
| chr3_146504144_146504452 | 0.41 |
Rpf1 |
ribosome production factor 1 homolog |
3926 |
0.15 |
| chr11_111269853_111270023 | 0.41 |
Gm11675 |
predicted gene 11675 |
6162 |
0.34 |
| chr10_107041161_107041312 | 0.40 |
Gm18040 |
predicted gene, 18040 |
18113 |
0.19 |
| chr2_58773911_58774203 | 0.40 |
Upp2 |
uridine phosphorylase 2 |
8732 |
0.21 |
| chr10_87872347_87872727 | 0.40 |
Igf1os |
insulin-like growth factor 1, opposite strand |
9156 |
0.2 |
| chr15_62529162_62529323 | 0.40 |
Gm41333 |
predicted gene, 41333 |
38733 |
0.21 |
| chr7_47035073_47035236 | 0.39 |
Gm45474 |
predicted gene 45474 |
15142 |
0.08 |
| chr6_140301064_140301256 | 0.39 |
Gm3961 |
predicted gene 3961 |
4704 |
0.2 |
| chr11_51727078_51727230 | 0.39 |
Gm25291 |
predicted gene, 25291 |
10768 |
0.14 |
| chr11_32651005_32651314 | 0.39 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
8255 |
0.23 |
| chr15_55186898_55187049 | 0.38 |
Deptor |
DEP domain containing MTOR-interacting protein |
53520 |
0.11 |
| chr1_164858611_164858762 | 0.38 |
Gm20743 |
predicted gene, 20743 |
4447 |
0.19 |
| chr13_34990551_34990817 | 0.38 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
1119 |
0.36 |
| chr9_20730600_20730850 | 0.38 |
Olfm2 |
olfactomedin 2 |
2496 |
0.23 |
| chr18_55793738_55793905 | 0.38 |
Gm8614 |
predicted gene 8614 |
45128 |
0.14 |
| chr19_45902358_45902683 | 0.37 |
Gm6813 |
predicted gene 6813 |
24394 |
0.14 |
| chr19_44394517_44394772 | 0.37 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12046 |
0.14 |
| chr10_86689548_86689699 | 0.37 |
Gm15344 |
predicted gene 15344 |
3472 |
0.09 |
| chr16_31432572_31432723 | 0.37 |
Gm15743 |
predicted gene 15743 |
2871 |
0.17 |
| chr2_132595408_132595654 | 0.37 |
AU019990 |
expressed sequence AU019990 |
2664 |
0.22 |
| chr1_88055296_88055515 | 0.37 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
6 |
0.94 |
| chr16_46623333_46623484 | 0.37 |
Gm17900 |
predicted gene, 17900 |
85573 |
0.1 |
| chr5_87019691_87019873 | 0.37 |
Gm18635 |
predicted gene, 18635 |
5622 |
0.12 |
| chr3_89147086_89147561 | 0.37 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
3891 |
0.08 |
| chr15_53297534_53297744 | 0.37 |
Ext1 |
exostosin glycosyltransferase 1 |
48020 |
0.18 |
| chr13_80895235_80895839 | 0.37 |
Arrdc3 |
arrestin domain containing 3 |
5019 |
0.18 |
| chr19_44027305_44027461 | 0.36 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
1816 |
0.28 |
| chr5_135367975_135368144 | 0.36 |
Nsun5 |
NOL1/NOP2/Sun domain family, member 5 |
1894 |
0.22 |
| chr5_33465562_33465766 | 0.36 |
Gm43851 |
predicted gene 43851 |
28200 |
0.15 |
| chr2_160848002_160848255 | 0.36 |
Gm11447 |
predicted gene 11447 |
2515 |
0.22 |
| chr2_4546547_4546716 | 0.36 |
Frmd4a |
FERM domain containing 4A |
13123 |
0.2 |
| chr9_103363870_103364326 | 0.36 |
Cdv3 |
carnitine deficiency-associated gene expressed in ventricle 3 |
799 |
0.56 |
| chr11_119048222_119048411 | 0.36 |
Cbx8 |
chromobox 8 |
7347 |
0.16 |
| chr9_122357372_122357607 | 0.35 |
Abhd5 |
abhydrolase domain containing 5 |
5700 |
0.15 |
| chr6_71999990_72000187 | 0.35 |
Gm26628 |
predicted gene, 26628 |
36313 |
0.1 |
| chr2_14596063_14596214 | 0.35 |
Cacnb2 |
calcium channel, voltage-dependent, beta 2 subunit |
6950 |
0.13 |
| chr16_93369696_93369958 | 0.35 |
Mir802 |
microRNA 802 |
107 |
0.95 |
| chr11_28702228_28702389 | 0.35 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
20744 |
0.15 |
| chr3_93589735_93589907 | 0.35 |
S100a10 |
S100 calcium binding protein A10 (calpactin) |
29094 |
0.09 |
| chr9_104246994_104247311 | 0.35 |
Dnajc13 |
DnaJ heat shock protein family (Hsp40) member C13 |
325 |
0.87 |
| chr10_115337191_115337351 | 0.34 |
Tmem19 |
transmembrane protein 19 |
11793 |
0.14 |
| chr7_66740464_66740653 | 0.34 |
Gm10623 |
predicted gene 10623 |
2754 |
0.19 |
| chr19_55110942_55111169 | 0.34 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
7964 |
0.2 |
| chr9_14061834_14061992 | 0.34 |
1700019J19Rik |
RIKEN cDNA 1700019J19 gene |
27707 |
0.17 |
| chr7_118715006_118715199 | 0.33 |
Ccp110 |
centriolar coiled coil protein 110 |
272 |
0.86 |
| chr14_60413760_60413920 | 0.33 |
Gm6066 |
predicted gene 6066 |
28139 |
0.17 |
| chr7_135720125_135720308 | 0.33 |
Mki67 |
antigen identified by monoclonal antibody Ki 67 |
3855 |
0.2 |
| chr3_144446211_144446394 | 0.33 |
Gm5857 |
predicted gene 5857 |
18630 |
0.18 |
| chr9_67824580_67824733 | 0.33 |
C2cd4a |
C2 calcium-dependent domain containing 4A |
7674 |
0.18 |
| chr18_39446995_39447226 | 0.33 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
40122 |
0.16 |
| chr19_3557401_3557552 | 0.33 |
Ppp6r3 |
protein phosphatase 6, regulatory subunit 3 |
18220 |
0.14 |
| chr6_118990185_118990336 | 0.33 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
89747 |
0.09 |
| chr3_18164974_18165125 | 0.33 |
Gm23686 |
predicted gene, 23686 |
12576 |
0.23 |
| chr4_116689870_116690150 | 0.33 |
Prdx1 |
peroxiredoxin 1 |
2698 |
0.15 |
| chr8_24445565_24445716 | 0.33 |
Gm44620 |
predicted gene 44620 |
1457 |
0.34 |
| chr5_90465453_90465660 | 0.32 |
Alb |
albumin |
2856 |
0.22 |
| chr10_69253303_69253474 | 0.32 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
12249 |
0.19 |
| chr3_136674419_136674771 | 0.32 |
Ppp3ca |
protein phosphatase 3, catalytic subunit, alpha isoform |
3825 |
0.27 |
| chr10_89573725_89573906 | 0.32 |
Gm48087 |
predicted gene, 48087 |
18096 |
0.17 |
| chr11_16817054_16817205 | 0.32 |
Egfros |
epidermal growth factor receptor, opposite strand |
13573 |
0.21 |
| chr14_30530847_30531018 | 0.32 |
Dcp1a |
decapping mRNA 1A |
7225 |
0.15 |
| chrX_60392083_60392234 | 0.32 |
Mir505 |
microRNA 505 |
2334 |
0.31 |
| chr3_109307549_109307748 | 0.32 |
Vav3 |
vav 3 oncogene |
33005 |
0.21 |
| chr15_53238274_53238425 | 0.32 |
Ext1 |
exostosin glycosyltransferase 1 |
42094 |
0.21 |
| chr12_117728620_117728794 | 0.32 |
Gm18955 |
predicted gene, 18955 |
4735 |
0.24 |
| chrX_95163511_95163805 | 0.32 |
Arhgef9 |
CDC42 guanine nucleotide exchange factor (GEF) 9 |
2167 |
0.36 |
| chr7_87371791_87371944 | 0.32 |
Tyr |
tyrosinase |
121525 |
0.05 |
| chr13_112316831_112317157 | 0.32 |
Ankrd55 |
ankyrin repeat domain 55 |
1460 |
0.39 |
| chr9_74791015_74791321 | 0.31 |
Gm22315 |
predicted gene, 22315 |
9098 |
0.19 |
| chr3_61217316_61217479 | 0.31 |
Gm37719 |
predicted gene, 37719 |
74049 |
0.1 |
| chr1_23913075_23913318 | 0.31 |
Smap1 |
small ArfGAP 1 |
3460 |
0.31 |
| chr4_135819951_135820102 | 0.31 |
Myom3 |
myomesin family, member 3 |
19382 |
0.11 |
| chr2_73491256_73491419 | 0.31 |
Wipf1 |
WAS/WASL interacting protein family, member 1 |
4868 |
0.19 |
| chr17_43858229_43858402 | 0.31 |
Rcan2 |
regulator of calcineurin 2 |
54312 |
0.15 |
| chr9_74319240_74319391 | 0.31 |
Gm24141 |
predicted gene, 24141 |
43295 |
0.15 |
| chr16_43408501_43408652 | 0.31 |
Gm15713 |
predicted gene 15713 |
11620 |
0.2 |
| chr18_65154932_65155123 | 0.31 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
707 |
0.74 |
| chr11_51753632_51753825 | 0.31 |
Sec24a |
Sec24 related gene family, member A (S. cerevisiae) |
2483 |
0.22 |
| chr1_67182326_67182537 | 0.31 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
59405 |
0.11 |
| chr17_53869978_53870142 | 0.31 |
Mrps36-ps1 |
mitichondrial ribosomal protein S36, pseudogene 1 |
3040 |
0.16 |
| chr16_43037670_43037825 | 0.31 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
82116 |
0.09 |
| chr19_14516462_14516647 | 0.31 |
Tle4 |
transducin-like enhancer of split 4 |
78985 |
0.11 |
| chr4_40698726_40698877 | 0.30 |
Aptx |
aprataxin |
3915 |
0.15 |
| chr2_82446048_82446229 | 0.30 |
A130030D18Rik |
RIKEN cDNA A130030D18 gene |
53936 |
0.14 |
| chr10_53398045_53398224 | 0.30 |
Gm36229 |
predicted gene, 36229 |
11748 |
0.13 |
| chr13_8847939_8848370 | 0.30 |
Wdr37 |
WD repeat domain 37 |
517 |
0.67 |
| chr1_163817675_163817827 | 0.30 |
Kifap3 |
kinesin-associated protein 3 |
38168 |
0.16 |
| chr15_3495710_3495861 | 0.30 |
Ghr |
growth hormone receptor |
24141 |
0.25 |
| chr18_12682851_12683167 | 0.30 |
Ttc39c |
tetratricopeptide repeat domain 39C |
6719 |
0.16 |
| chr2_18048778_18049088 | 0.30 |
Skida1 |
SKI/DACH domain containing 1 |
98 |
0.94 |
| chr18_39827979_39828276 | 0.30 |
Pabpc2 |
poly(A) binding protein, cytoplasmic 2 |
54630 |
0.14 |
| chr6_65569857_65570008 | 0.30 |
Tnip3 |
TNFAIP3 interacting protein 3 |
20466 |
0.22 |
| chr2_5939653_5939804 | 0.30 |
Dhtkd1 |
dehydrogenase E1 and transketolase domain containing 1 |
3015 |
0.22 |
| chr2_113795317_113795493 | 0.30 |
Scg5 |
secretogranin V |
33357 |
0.14 |
| chr10_99172107_99172427 | 0.30 |
Poc1b |
POC1 centriolar protein B |
983 |
0.45 |
| chr1_107939441_107939624 | 0.30 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
2823 |
0.25 |
| chr5_87455158_87455319 | 0.30 |
Ugt2a2 |
UDP glucuronosyltransferase 2 family, polypeptide A2 |
27020 |
0.09 |
| chr13_5804589_5804740 | 0.30 |
Gm26043 |
predicted gene, 26043 |
17194 |
0.19 |
| chr17_54414428_54414603 | 0.30 |
Trp53-ps |
transformation related protein 53, pseudogene |
5165 |
0.22 |
| chr16_21779191_21779393 | 0.30 |
Gm24898 |
predicted gene, 24898 |
5179 |
0.14 |
| chr6_134743647_134743846 | 0.29 |
Dusp16 |
dual specificity phosphatase 16 |
80 |
0.96 |
| chr10_38136267_38136418 | 0.29 |
Gm31378 |
predicted gene, 31378 |
14281 |
0.19 |
| chr1_134567570_134567757 | 0.29 |
Kdm5b |
lysine (K)-specific demethylase 5B |
7456 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.3 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.5 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.0 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.2 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0045713 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.0 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.0 | GO:0072221 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |