Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
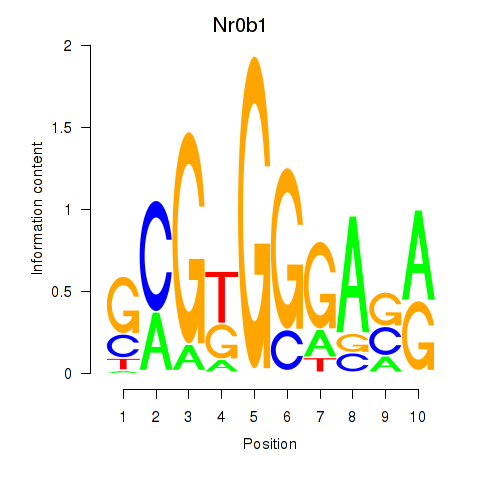
Results for Nr0b1
Z-value: 2.65
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.4 | nuclear receptor subfamily 0, group B, member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_86216560_86216728 | Nr0b1 | 24880 | 0.154148 | -0.45 | 3.7e-01 | Click! |
Activity of the Nr0b1 motif across conditions
Conditions sorted by the z-value of the Nr0b1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
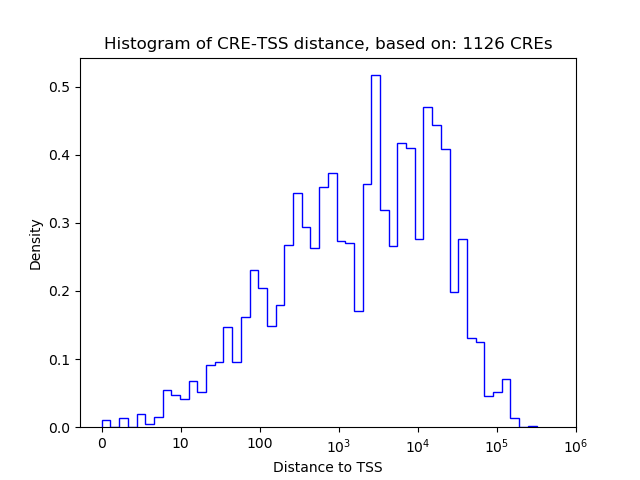
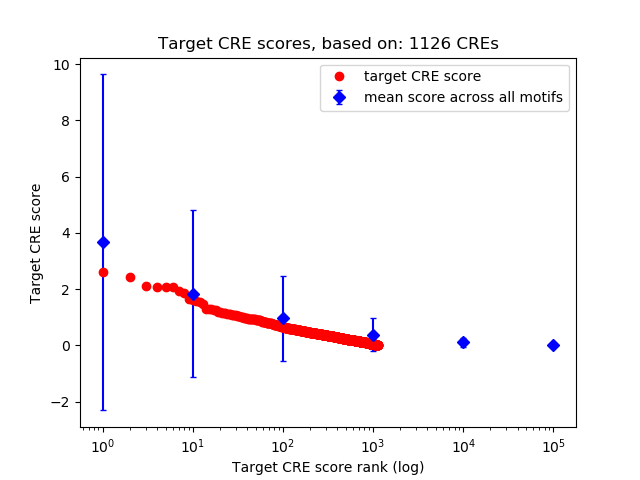
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_106640189_106640344 | 2.61 |
Cdh1 |
cadherin 1 |
36123 |
0.13 |
| chr1_74331641_74331792 | 2.42 |
Pnkd |
paroxysmal nonkinesiogenic dyskinesia |
894 |
0.38 |
| chr11_107130421_107130628 | 2.10 |
Bptf |
bromodomain PHD finger transcription factor |
1398 |
0.36 |
| chr17_56583583_56583743 | 2.07 |
Safb2 |
scaffold attachment factor B2 |
617 |
0.5 |
| chr16_95824094_95824245 | 2.07 |
2810404F17Rik |
RIKEN cDNA 2810404F17 gene |
15852 |
0.17 |
| chr1_52388262_52388419 | 2.07 |
Gm23460 |
predicted gene, 23460 |
56204 |
0.11 |
| chr12_25132144_25132315 | 1.92 |
Gm17746 |
predicted gene, 17746 |
3055 |
0.22 |
| chr7_28862945_28863096 | 1.85 |
Lgals7 |
lectin, galactose binding, soluble 7 |
833 |
0.39 |
| chr5_137398768_137398919 | 1.63 |
Zan |
zonadhesin |
2895 |
0.15 |
| chr6_124694855_124695031 | 1.62 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
6476 |
0.07 |
| chr14_54417307_54417458 | 1.59 |
Slc7a7 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
297 |
0.79 |
| chr14_54897515_54897687 | 1.55 |
Pabpn1 |
poly(A) binding protein, nuclear 1 |
2768 |
0.09 |
| chr8_119417171_119417322 | 1.47 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
16878 |
0.13 |
| chr2_94273299_94273450 | 1.31 |
Mir670hg |
MIR670 host gene (non-protein coding) |
8456 |
0.15 |
| chr11_51832795_51832987 | 1.28 |
Jade2 |
jade family PHD finger 2 |
24234 |
0.15 |
| chrX_140524377_140524528 | 1.28 |
Tsc22d3 |
TSC22 domain family, member 3 |
18216 |
0.18 |
| chr12_104798236_104798545 | 1.26 |
Clmn |
calmin |
17519 |
0.19 |
| chr1_170489592_170489765 | 1.24 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
34764 |
0.18 |
| chr9_32718091_32718269 | 1.19 |
Gm27240 |
predicted gene 27240 |
7009 |
0.2 |
| chr3_88163993_88164144 | 1.18 |
Mef2d |
myocyte enhancer factor 2D |
1108 |
0.31 |
| chr14_101652712_101652891 | 1.15 |
Uchl3 |
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
1166 |
0.44 |
| chr6_125575019_125575372 | 1.14 |
Vwf |
Von Willebrand factor |
8944 |
0.21 |
| chr2_163166224_163166432 | 1.14 |
Gm14254 |
predicted gene 14254 |
10222 |
0.16 |
| chr14_18678485_18678659 | 1.11 |
Ube2e2 |
ubiquitin-conjugating enzyme E2E 2 |
13422 |
0.25 |
| chr2_148685098_148685392 | 1.11 |
Gzf1 |
GDNF-inducible zinc finger protein 1 |
633 |
0.66 |
| chr9_107908957_107909108 | 1.10 |
Mst1r |
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
2125 |
0.13 |
| chr9_98429359_98429510 | 1.09 |
Rbp1 |
retinol binding protein 1, cellular |
6473 |
0.22 |
| chr5_33694927_33695093 | 1.08 |
Gm42965 |
predicted gene 42965 |
12036 |
0.1 |
| chr17_27636792_27636990 | 1.07 |
Rps10 |
ribosomal protein S10 |
222 |
0.82 |
| chr5_75046920_75047087 | 1.06 |
Gm18593 |
predicted gene, 18593 |
1476 |
0.27 |
| chr12_105784205_105784562 | 1.05 |
Papola |
poly (A) polymerase alpha |
311 |
0.9 |
| chr7_113219504_113219670 | 1.05 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
12030 |
0.21 |
| chr1_165772571_165772722 | 1.03 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
3170 |
0.12 |
| chr5_120648695_120648846 | 1.01 |
Rasal1 |
RAS protein activator like 1 (GAP1 like) |
42 |
0.91 |
| chr2_5458815_5458972 | 0.99 |
Gm13216 |
predicted gene 13216 |
144678 |
0.04 |
| chr15_36960858_36961018 | 0.99 |
Gm34590 |
predicted gene, 34590 |
22074 |
0.14 |
| chr6_67088817_67089020 | 0.98 |
E230016M11Rik |
RIKEN cDNA E230016M11 gene |
12158 |
0.14 |
| chr4_41546048_41546413 | 0.98 |
Fam219a |
family with sequence similarity 219, member A |
21460 |
0.09 |
| chr9_63662886_63663067 | 0.97 |
Smad3 |
SMAD family member 3 |
3571 |
0.25 |
| chr9_70243258_70243430 | 0.97 |
Myo1e |
myosin IE |
35976 |
0.17 |
| chr1_72833162_72833313 | 0.95 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
7915 |
0.22 |
| chr5_63964660_63964830 | 0.95 |
Rell1 |
RELT-like 1 |
4078 |
0.19 |
| chr11_67038930_67039086 | 0.94 |
Adprm |
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
1519 |
0.29 |
| chr5_134571935_134572111 | 0.94 |
Mir7228 |
microRNA 7228 |
53 |
0.95 |
| chr9_66802090_66802293 | 0.94 |
BC050972 |
cDNA sequence BC050972 |
368 |
0.78 |
| chr7_127803770_127804052 | 0.93 |
9430064I24Rik |
RIKEN cDNA 9430064I24 gene |
1149 |
0.22 |
| chr9_121909086_121909237 | 0.93 |
Ackr2 |
atypical chemokine receptor 2 |
2897 |
0.12 |
| chr7_31128675_31128826 | 0.93 |
Scn1b |
sodium channel, voltage-gated, type I, beta |
1747 |
0.19 |
| chr1_72014626_72014994 | 0.92 |
4933417E11Rik |
RIKEN cDNA 4933417E11 gene |
9615 |
0.16 |
| chr7_128398804_128398955 | 0.92 |
Rgs10 |
regulator of G-protein signalling 10 |
5194 |
0.13 |
| chr15_59717023_59717209 | 0.91 |
Gm20150 |
predicted gene, 20150 |
45003 |
0.15 |
| chr9_65084518_65084694 | 0.91 |
Dpp8 |
dipeptidylpeptidase 8 |
8858 |
0.14 |
| chr11_119035192_119035358 | 0.90 |
Cbx8 |
chromobox 8 |
5556 |
0.17 |
| chr8_67948591_67948742 | 0.90 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
3213 |
0.28 |
| chr14_30974299_30974450 | 0.89 |
Nek4 |
NIMA (never in mitosis gene a)-related expressed kinase 4 |
17459 |
0.08 |
| chr3_81427554_81427721 | 0.89 |
Gm37300 |
predicted gene, 37300 |
22065 |
0.27 |
| chr15_62079300_62079458 | 0.86 |
Pvt1 |
Pvt1 oncogene |
19650 |
0.25 |
| chr14_30579376_30579527 | 0.86 |
Mir3076 |
microRNA 3076 |
7302 |
0.14 |
| chr10_84576177_84576339 | 0.85 |
Tcp11l2 |
t-complex 11 (mouse) like 2 |
368 |
0.81 |
| chr2_128387976_128388132 | 0.84 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
14911 |
0.19 |
| chr18_76301148_76301315 | 0.84 |
Gm50360 |
predicted gene, 50360 |
20593 |
0.19 |
| chr2_33130281_33130432 | 0.84 |
Garnl3 |
GTPase activating RANGAP domain-like 3 |
252 |
0.91 |
| chr16_31576793_31576977 | 0.83 |
Gm34256 |
predicted gene, 34256 |
16335 |
0.16 |
| chr3_85821987_85822152 | 0.83 |
Fam160a1 |
family with sequence similarity 160, member A1 |
4778 |
0.2 |
| chr18_12191587_12191744 | 0.82 |
Gm15956 |
predicted gene 15956 |
2103 |
0.2 |
| chr5_9038233_9038384 | 0.80 |
Gm40264 |
predicted gene, 40264 |
3184 |
0.2 |
| chr19_42612808_42613017 | 0.80 |
Loxl4 |
lysyl oxidase-like 4 |
99 |
0.97 |
| chr19_42602482_42602861 | 0.80 |
Loxl4 |
lysyl oxidase-like 4 |
650 |
0.71 |
| chr1_131976711_131976867 | 0.80 |
Slc45a3 |
solute carrier family 45, member 3 |
70 |
0.96 |
| chr8_10899350_10899661 | 0.79 |
4833411C07Rik |
RIKEN cDNA 4833411C07 gene |
417 |
0.64 |
| chr2_38379337_38379528 | 0.78 |
Gm13584 |
predicted gene 13584 |
15452 |
0.14 |
| chr9_43545212_43545661 | 0.78 |
Gm36855 |
predicted gene, 36855 |
24432 |
0.17 |
| chr5_52996866_52997026 | 0.78 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
2510 |
0.23 |
| chr17_31569080_31569361 | 0.78 |
Gm50107 |
predicted gene, 50107 |
2057 |
0.13 |
| chr11_109850472_109850623 | 0.77 |
1700023C21Rik |
RIKEN cDNA 1700023C21 gene |
4669 |
0.21 |
| chr9_66733466_66733631 | 0.76 |
Car12 |
carbonic anhydrase 12 |
14162 |
0.17 |
| chr4_131939568_131939730 | 0.76 |
Epb41 |
erythrocyte membrane protein band 4.1 |
2262 |
0.18 |
| chr17_84877556_84877740 | 0.75 |
Gm49982 |
predicted gene, 49982 |
24921 |
0.14 |
| chr16_11066984_11067173 | 0.75 |
Snn |
stannin |
748 |
0.38 |
| chr10_63410268_63410432 | 0.73 |
Gm7530 |
predicted gene 7530 |
2649 |
0.19 |
| chr12_103820499_103820882 | 0.73 |
Gm8893 |
predicted gene 8893 |
1553 |
0.23 |
| chr15_97598448_97598613 | 0.73 |
Gm49506 |
predicted gene, 49506 |
40240 |
0.16 |
| chr1_119292285_119292453 | 0.73 |
Gm29456 |
predicted gene 29456 |
34464 |
0.14 |
| chr17_25472162_25472329 | 0.72 |
Tekt4 |
tektin 4 |
648 |
0.56 |
| chr11_57986258_57986432 | 0.72 |
Gm12249 |
predicted gene 12249 |
13211 |
0.15 |
| chr18_67933368_67933544 | 0.72 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
199 |
0.95 |
| chr2_43469286_43469452 | 0.71 |
Gm13464 |
predicted gene 13464 |
50029 |
0.18 |
| chr16_21779191_21779393 | 0.71 |
Gm24898 |
predicted gene, 24898 |
5179 |
0.14 |
| chr16_30068818_30068984 | 0.70 |
Hes1 |
hes family bHLH transcription factor 1 |
2563 |
0.23 |
| chr2_75612978_75613138 | 0.70 |
Gm13655 |
predicted gene 13655 |
20324 |
0.15 |
| chr4_124215186_124215370 | 0.70 |
Gm37667 |
predicted gene, 37667 |
63899 |
0.1 |
| chr5_122951419_122951624 | 0.69 |
Kdm2b |
lysine (K)-specific demethylase 2B |
469 |
0.74 |
| chr10_126997803_126998005 | 0.67 |
Mir546 |
microRNA 546 |
536 |
0.51 |
| chr2_166092956_166093107 | 0.67 |
Sulf2 |
sulfatase 2 |
14912 |
0.17 |
| chr9_70138613_70138777 | 0.67 |
Fam81a |
family with sequence similarity 81, member A |
2883 |
0.24 |
| chr11_102143113_102143282 | 0.66 |
Nags |
N-acetylglutamate synthase |
2316 |
0.13 |
| chr19_43525218_43525376 | 0.66 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
692 |
0.55 |
| chr5_140699850_140700001 | 0.66 |
Iqce |
IQ motif containing E |
2044 |
0.26 |
| chr15_99724752_99724954 | 0.66 |
Gm16537 |
predicted gene 16537 |
379 |
0.58 |
| chr6_39205376_39205554 | 0.65 |
Kdm7a |
lysine (K)-specific demethylase 7A |
1324 |
0.32 |
| chr9_121376171_121376353 | 0.64 |
Trak1 |
trafficking protein, kinesin binding 1 |
6532 |
0.19 |
| chr15_102014758_102014925 | 0.64 |
Krt8 |
keratin 8 |
10359 |
0.11 |
| chr1_72824439_72824598 | 0.64 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
15 |
0.98 |
| chr3_115789167_115789324 | 0.64 |
Gm9889 |
predicted gene 9889 |
74095 |
0.08 |
| chr11_115159020_115159171 | 0.63 |
Slc9a3r1 |
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
4246 |
0.13 |
| chr11_75444470_75444637 | 0.63 |
Serpinf2 |
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
4962 |
0.09 |
| chr18_67201855_67202117 | 0.63 |
Chmp1b |
charged multivesicular body protein 1B |
3380 |
0.17 |
| chr2_25094968_25095119 | 0.63 |
Noxa1 |
NADPH oxidase activator 1 |
88 |
0.93 |
| chr8_128484955_128485414 | 0.62 |
Nrp1 |
neuropilin 1 |
125787 |
0.05 |
| chr14_55590995_55591169 | 0.61 |
Psme2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
5 |
0.87 |
| chr9_57696029_57696203 | 0.61 |
Cyp1a1 |
cytochrome P450, family 1, subfamily a, polypeptide 1 |
1497 |
0.28 |
| chr15_102150406_102150578 | 0.61 |
Soat2 |
sterol O-acyltransferase 2 |
34 |
0.95 |
| chr13_101694986_101695178 | 0.60 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
482 |
0.84 |
| chr2_27746034_27746215 | 0.60 |
Rxra |
retinoid X receptor alpha |
5923 |
0.28 |
| chr1_134513989_134514156 | 0.60 |
Gm15454 |
predicted gene 15454 |
4009 |
0.12 |
| chr13_53125792_53125963 | 0.60 |
Gm48336 |
predicted gene, 48336 |
96959 |
0.07 |
| chr19_43896493_43896674 | 0.60 |
Dnmbp |
dynamin binding protein |
5892 |
0.15 |
| chr4_144956318_144956685 | 0.60 |
Gm38074 |
predicted gene, 38074 |
2347 |
0.3 |
| chr1_133085141_133085309 | 0.59 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
16646 |
0.13 |
| chr16_30596865_30597035 | 0.59 |
Fam43a |
family with sequence similarity 43, member A |
2773 |
0.27 |
| chr18_64349943_64350119 | 0.59 |
Onecut2 |
one cut domain, family member 2 |
10011 |
0.17 |
| chr10_8195740_8196032 | 0.59 |
Gm30906 |
predicted gene, 30906 |
19755 |
0.25 |
| chr3_152402367_152402529 | 0.59 |
Zzz3 |
zinc finger, ZZ domain containing 3 |
5754 |
0.12 |
| chr4_154497558_154497782 | 0.59 |
Prdm16 |
PR domain containing 16 |
31095 |
0.16 |
| chr6_94696254_94696442 | 0.59 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
3789 |
0.28 |
| chr13_107798210_107798399 | 0.58 |
Gm22118 |
predicted gene, 22118 |
30976 |
0.19 |
| chr4_144901641_144901842 | 0.58 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
8522 |
0.21 |
| chr2_27676858_27677019 | 0.58 |
Rxra |
retinoid X receptor alpha |
263 |
0.94 |
| chr11_113063139_113063329 | 0.58 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
109843 |
0.07 |
| chr7_12978093_12978257 | 0.58 |
Zfp446 |
zinc finger protein 446 |
63 |
0.93 |
| chr18_35034038_35034225 | 0.58 |
Gm36037 |
predicted gene, 36037 |
51161 |
0.09 |
| chr6_113038436_113038611 | 0.57 |
Gm23244 |
predicted gene, 23244 |
2589 |
0.16 |
| chr16_77360217_77360390 | 0.57 |
Gm37466 |
predicted gene, 37466 |
21647 |
0.13 |
| chr15_85808535_85808715 | 0.56 |
Cdpf1 |
cysteine rich, DPF motif domain containing 1 |
2465 |
0.2 |
| chr12_27826295_27826490 | 0.56 |
Gm45941 |
predicted gene, 45941 |
119650 |
0.06 |
| chr15_36471981_36472313 | 0.56 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr17_46030522_46030717 | 0.56 |
Vegfa |
vascular endothelial growth factor A |
737 |
0.63 |
| chr12_25092894_25093391 | 0.56 |
Id2 |
inhibitor of DNA binding 2 |
2945 |
0.22 |
| chrX_48193537_48193988 | 0.56 |
Zdhhc9 |
zinc finger, DHHC domain containing 9 |
14566 |
0.16 |
| chr11_97138815_97139024 | 0.55 |
Mir7235 |
microRNA 7235 |
7500 |
0.09 |
| chrX_12159268_12159419 | 0.55 |
Bcor |
BCL6 interacting corepressor |
1003 |
0.62 |
| chr3_95905207_95905371 | 0.55 |
Car14 |
carbonic anhydrase 14 |
598 |
0.51 |
| chr3_122482421_122482595 | 0.55 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
1215 |
0.39 |
| chr12_103856410_103857076 | 0.54 |
Serpina1a |
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
6808 |
0.1 |
| chr19_30092786_30092953 | 0.54 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
908 |
0.62 |
| chr9_50604082_50604278 | 0.54 |
Timm8b |
translocase of inner mitochondrial membrane 8B |
270 |
0.56 |
| chr8_33803402_33803767 | 0.54 |
Rbpms |
RNA binding protein gene with multiple splicing |
3215 |
0.2 |
| chr4_44979379_44979530 | 0.54 |
Gm12679 |
predicted gene 12679 |
1135 |
0.32 |
| chr19_6242419_6242590 | 0.54 |
Atg2a |
autophagy related 2A |
836 |
0.36 |
| chr19_47313567_47313744 | 0.53 |
Sh3pxd2a |
SH3 and PX domains 2A |
1096 |
0.49 |
| chr11_115535817_115535982 | 0.53 |
Sumo2 |
small ubiquitin-like modifier 2 |
82 |
0.94 |
| chr12_105039919_105040249 | 0.53 |
Glrx5 |
glutaredoxin 5 |
4868 |
0.11 |
| chr2_80581456_80581770 | 0.53 |
Nckap1 |
NCK-associated protein 1 |
233 |
0.89 |
| chr10_25595963_25596295 | 0.53 |
Gm36908 |
predicted gene, 36908 |
25434 |
0.15 |
| chr13_73723026_73723388 | 0.53 |
Slc12a7 |
solute carrier family 12, member 7 |
9887 |
0.15 |
| chr12_83582014_83582393 | 0.52 |
Zfyve1 |
zinc finger, FYVE domain containing 1 |
13484 |
0.14 |
| chr2_167888871_167889131 | 0.52 |
Gm14319 |
predicted gene 14319 |
30416 |
0.13 |
| chrX_70792837_70793020 | 0.52 |
Gm8410 |
predicted gene 8410 |
22061 |
0.21 |
| chr9_90260181_90260562 | 0.52 |
Tbc1d2b |
TBC1 domain family, member 2B |
4444 |
0.2 |
| chr7_89417848_89418045 | 0.52 |
AI314278 |
expressed sequence AI314278 |
8360 |
0.14 |
| chr12_100200505_100200696 | 0.51 |
Calm1 |
calmodulin 1 |
456 |
0.75 |
| chr2_165304914_165305101 | 0.51 |
Elmo2 |
engulfment and cell motility 2 |
6635 |
0.15 |
| chr12_99468179_99468384 | 0.51 |
Foxn3 |
forkhead box N3 |
18184 |
0.18 |
| chr5_66080941_66081107 | 0.51 |
Rbm47 |
RNA binding motif protein 47 |
24 |
0.96 |
| chr1_67124558_67125159 | 0.50 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
1832 |
0.42 |
| chr10_95678111_95678450 | 0.50 |
Anapc15-ps |
anaphase promoting complex C subunit 15, pseudogene |
4829 |
0.14 |
| chr14_75753944_75754135 | 0.50 |
Cog3 |
component of oligomeric golgi complex 3 |
519 |
0.74 |
| chr2_4156676_4156835 | 0.50 |
Frmd4a |
FERM domain containing 4A |
3883 |
0.14 |
| chr5_139358617_139359046 | 0.50 |
Cyp2w1 |
cytochrome P450, family 2, subfamily w, polypeptide 1 |
4255 |
0.13 |
| chr5_125326284_125326660 | 0.50 |
Scarb1 |
scavenger receptor class B, member 1 |
205 |
0.92 |
| chr6_136210853_136211004 | 0.50 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
37417 |
0.15 |
| chr14_62492589_62492750 | 0.50 |
Gm4131 |
predicted gene 4131 |
11514 |
0.13 |
| chr7_71468093_71468244 | 0.50 |
Gm29328 |
predicted gene 29328 |
97835 |
0.07 |
| chr17_29078234_29078394 | 0.49 |
Trp53cor1 |
tumor protein p53 pathway corepressor 1 |
865 |
0.39 |
| chr8_26698632_26698820 | 0.49 |
Gm32098 |
predicted gene, 32098 |
30420 |
0.14 |
| chr11_75053082_75053244 | 0.49 |
Gm12335 |
predicted gene 12335 |
13599 |
0.12 |
| chr14_62245516_62245667 | 0.49 |
Gm26969 |
predicted gene, 26969 |
41239 |
0.13 |
| chr7_88430182_88430353 | 0.49 |
Rab38 |
RAB38, member RAS oncogene family |
6 |
0.98 |
| chr10_118867636_118867819 | 0.49 |
Dyrk2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
572 |
0.66 |
| chr14_34502578_34502742 | 0.49 |
Bmpr1a |
bone morphogenetic protein receptor, type 1A |
27 |
0.94 |
| chr11_45852561_45852757 | 0.49 |
Clint1 |
clathrin interactor 1 |
695 |
0.62 |
| chr7_139247418_139247589 | 0.49 |
Pwwp2b |
PWWP domain containing 2B |
979 |
0.48 |
| chr4_55015194_55015412 | 0.49 |
Zfp462 |
zinc finger protein 462 |
3823 |
0.31 |
| chr17_65950804_65950984 | 0.49 |
Twsg1 |
twisted gastrulation BMP signaling modulator 1 |
221 |
0.91 |
| chr16_24056611_24056770 | 0.49 |
Gm49518 |
predicted gene, 49518 |
16784 |
0.15 |
| chr14_31594823_31594990 | 0.48 |
Colq |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
3563 |
0.19 |
| chr3_89422230_89422381 | 0.48 |
Shc1 |
src homology 2 domain-containing transforming protein C1 |
663 |
0.44 |
| chr16_48258588_48258739 | 0.48 |
Gm49770 |
predicted gene, 49770 |
3151 |
0.23 |
| chr14_27325762_27325922 | 0.47 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
10224 |
0.22 |
| chr2_29750814_29750965 | 0.47 |
Gm13420 |
predicted gene 13420 |
796 |
0.47 |
| chr9_21723482_21723643 | 0.47 |
Ldlr |
low density lipoprotein receptor |
37 |
0.58 |
| chr14_46017259_46017572 | 0.47 |
Gm6580 |
predicted gene 6580 |
10013 |
0.17 |
| chr8_13200336_13200487 | 0.47 |
Grtp1 |
GH regulated TBC protein 1 |
124 |
0.72 |
| chr17_15375757_15376124 | 0.47 |
Dll1 |
delta like canonical Notch ligand 1 |
36 |
0.97 |
| chr3_83029657_83029978 | 0.47 |
Fga |
fibrinogen alpha chain |
3602 |
0.17 |
| chr4_137481328_137481504 | 0.47 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
12613 |
0.13 |
| chr14_54417687_54417851 | 0.46 |
Slc7a7 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
11 |
0.95 |
| chr3_82998880_82999031 | 0.46 |
Gm30097 |
predicted gene, 30097 |
3370 |
0.19 |
| chr3_145988519_145988702 | 0.46 |
Syde2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
43 |
0.97 |
| chr5_74315924_74316091 | 0.46 |
Gm15981 |
predicted gene 15981 |
1027 |
0.5 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 | 0.8 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.2 | 1.4 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.2 | 0.5 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.2 | 0.9 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.5 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 | 0.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.2 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.5 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.2 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.5 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.2 | 0.5 | GO:0048370 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.1 | 0.4 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.5 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.4 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.3 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.3 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.1 | 0.4 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.3 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.1 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.3 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.1 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.8 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 0.2 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.1 | 0.2 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.1 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.9 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.6 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.1 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.1 | 0.4 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.2 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 1.0 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.6 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.1 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.2 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.2 | GO:1901859 | negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 0.6 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.1 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.8 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.3 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.8 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:1901838 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.3 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 1.0 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.7 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0035912 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:1900164 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.0 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:1902566 | regulation of eosinophil activation(GO:1902566) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) |
| 0.0 | 0.6 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.0 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.5 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.2 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.0 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.0 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.0 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0061623 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.0 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) |
| 0.0 | 0.5 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.0 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 0.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 0.8 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.6 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 1.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.5 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 0.5 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.3 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.1 | 0.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0043723 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 1.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0052622 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |