Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
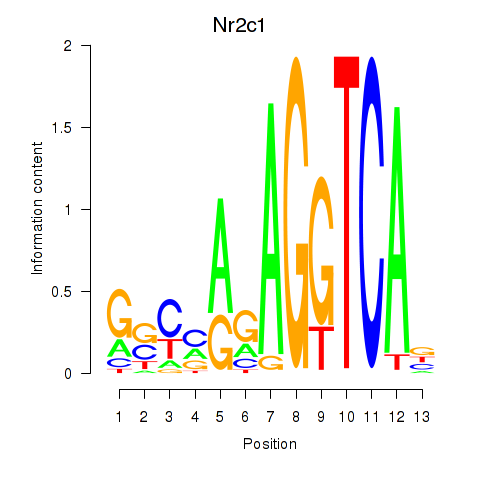
Results for Nr2c1
Z-value: 1.80
Transcription factors associated with Nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c1
|
ENSMUSG00000005897.8 | nuclear receptor subfamily 2, group C, member 1 |
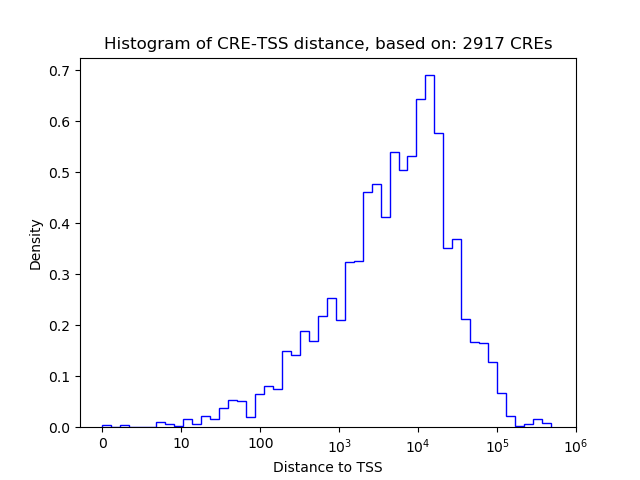
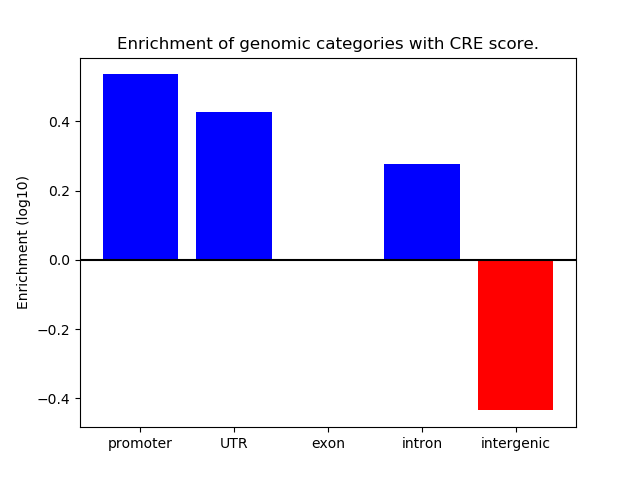
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_94159224_94159471 | Nr2c1 | 3061 | 0.190210 | 0.75 | 8.8e-02 | Click! |
| chr10_94160795_94160946 | Nr2c1 | 4584 | 0.161402 | -0.74 | 9.5e-02 | Click! |
| chr10_94162152_94162401 | Nr2c1 | 5990 | 0.150969 | -0.73 | 9.8e-02 | Click! |
| chr10_94159643_94159802 | Nr2c1 | 3436 | 0.179798 | 0.67 | 1.5e-01 | Click! |
| chr10_94150437_94150761 | Nr2c1 | 2180 | 0.232468 | 0.62 | 1.9e-01 | Click! |
Activity of the Nr2c1 motif across conditions
Conditions sorted by the z-value of the Nr2c1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
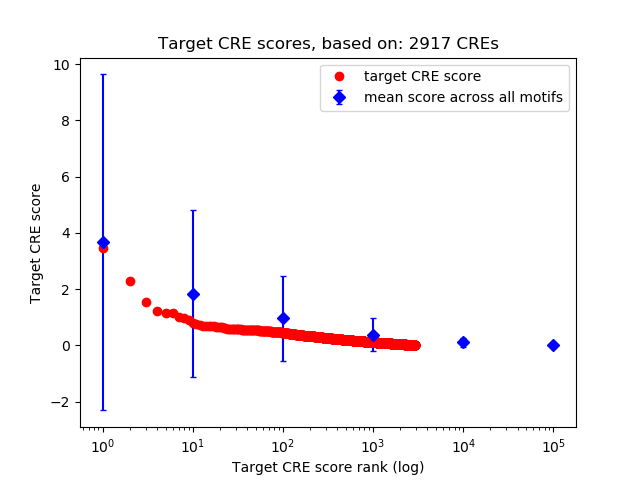
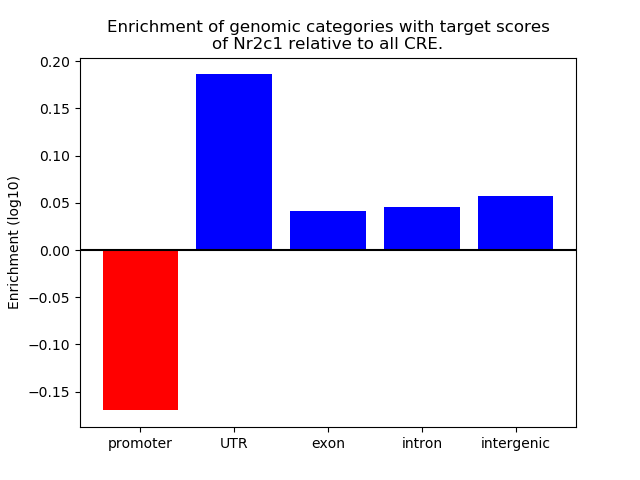
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_98301768_98301944 | 3.47 |
Gm20644 |
predicted gene 20644 |
16474 |
0.09 |
| chr16_32870319_32870472 | 2.30 |
Rubcn |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
2029 |
0.24 |
| chr3_52646413_52646568 | 1.52 |
Gm10293 |
predicted pseudogene 10293 |
33655 |
0.18 |
| chr8_125976536_125976742 | 1.23 |
Kcnk1 |
potassium channel, subfamily K, member 1 |
18531 |
0.19 |
| chr4_144973561_144973712 | 1.14 |
Vps13d |
vacuolar protein sorting 13D |
10325 |
0.2 |
| chr17_28028156_28028307 | 1.14 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
12544 |
0.11 |
| chr8_24160713_24160865 | 1.02 |
A730045E13Rik |
RIKEN cDNA A730045E13 gene |
13115 |
0.25 |
| chr5_117089265_117089549 | 0.96 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
9404 |
0.15 |
| chr5_144787968_144788267 | 0.89 |
Trrap |
transformation/transcription domain-associated protein |
2592 |
0.24 |
| chr18_9489453_9489931 | 0.81 |
Gm7527 |
predicted gene 7527 |
5929 |
0.17 |
| chr13_24936133_24936311 | 0.75 |
9330162012Rik |
cDNA RIKEN 9330162012 gene |
1179 |
0.31 |
| chr17_84147026_84147177 | 0.71 |
Gm19696 |
predicted gene, 19696 |
7260 |
0.17 |
| chr6_32962622_32962773 | 0.70 |
Chchd3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
65626 |
0.11 |
| chr6_139901873_139902024 | 0.70 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
16035 |
0.22 |
| chr14_11564616_11565059 | 0.69 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
11256 |
0.22 |
| chr7_28543442_28543608 | 0.68 |
Sycn |
syncollin |
2640 |
0.11 |
| chr1_52798933_52799346 | 0.67 |
Inpp1 |
inositol polyphosphate-1-phosphatase |
349 |
0.87 |
| chr4_59262947_59263121 | 0.67 |
Gm12596 |
predicted gene 12596 |
2983 |
0.26 |
| chr11_95778891_95779340 | 0.67 |
Polr2k-ps |
polymerase (RNA) II (DNA directed) polypeptide K, pseudogene |
17624 |
0.11 |
| chr6_99271808_99271987 | 0.66 |
Foxp1 |
forkhead box P1 |
5365 |
0.31 |
| chr4_125440047_125440198 | 0.64 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
50578 |
0.13 |
| chr1_133127295_133127631 | 0.63 |
Ppp1r15b |
protein phosphatase 1, regulatory subunit 15B |
3680 |
0.18 |
| chr17_33932704_33933052 | 0.62 |
Rgl2 |
ral guanine nucleotide dissociation stimulator-like 2 |
747 |
0.27 |
| chr12_8039741_8039902 | 0.59 |
Apob |
apolipoprotein B |
27462 |
0.2 |
| chr6_90499410_90499561 | 0.59 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
13058 |
0.1 |
| chr9_110456029_110456203 | 0.59 |
Klhl18 |
kelch-like 18 |
715 |
0.58 |
| chr9_48749463_48749719 | 0.59 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
86354 |
0.08 |
| chr14_30929998_30930386 | 0.59 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
6432 |
0.11 |
| chr18_9487035_9487277 | 0.59 |
Gm7527 |
predicted gene 7527 |
8465 |
0.16 |
| chr12_7751611_7751762 | 0.58 |
Gm32828 |
predicted gene, 32828 |
108005 |
0.06 |
| chr3_31136174_31136377 | 0.58 |
Cldn11 |
claudin 11 |
13645 |
0.2 |
| chr12_29257874_29258025 | 0.57 |
Gm6989 |
predicted gene 6989 |
59619 |
0.14 |
| chr14_21422563_21422878 | 0.57 |
Gm25864 |
predicted gene, 25864 |
27754 |
0.17 |
| chr11_117310188_117310356 | 0.57 |
Septin9 |
septin 9 |
2118 |
0.32 |
| chr13_57573617_57573793 | 0.56 |
Gm48176 |
predicted gene, 48176 |
297027 |
0.01 |
| chr10_92278112_92278274 | 0.56 |
Gm47124 |
predicted gene, 47124 |
19603 |
0.2 |
| chr6_72592358_72592612 | 0.56 |
Elmod3 |
ELMO/CED-12 domain containing 3 |
2536 |
0.14 |
| chr2_32416589_32417000 | 0.56 |
Slc25a25 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
223 |
0.86 |
| chr17_14988060_14988226 | 0.56 |
9030025P20Rik |
RIKEN cDNA 9030025P20 gene |
8644 |
0.11 |
| chr17_10878758_10878909 | 0.55 |
Gm3145 |
predicted gene 3145 |
20162 |
0.19 |
| chr12_75599874_75600059 | 0.55 |
Ppp2r5e |
protein phosphatase 2, regulatory subunit B', epsilon |
3721 |
0.22 |
| chr10_75033725_75033908 | 0.55 |
Gm40692 |
predicted gene, 40692 |
529 |
0.63 |
| chr14_25295396_25295547 | 0.55 |
Gm26660 |
predicted gene, 26660 |
83164 |
0.08 |
| chr11_60844761_60844937 | 0.55 |
Dhrs7b |
dehydrogenase/reductase (SDR family) member 7B |
14170 |
0.09 |
| chr10_42194110_42194261 | 0.55 |
Foxo3 |
forkhead box O3 |
64181 |
0.12 |
| chr6_6183421_6183599 | 0.54 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
7809 |
0.26 |
| chr8_70717164_70717873 | 0.54 |
Gm3336 |
predicted gene 3336 |
1025 |
0.27 |
| chr5_93296317_93296483 | 0.54 |
Ccng2 |
cyclin G2 |
27692 |
0.16 |
| chr6_6204143_6204472 | 0.54 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
12811 |
0.23 |
| chr11_23702943_23703137 | 0.53 |
Pus10 |
pseudouridylate synthase 10 |
14442 |
0.16 |
| chr16_10644168_10644607 | 0.53 |
Clec16a |
C-type lectin domain family 16, member A |
14327 |
0.18 |
| chr7_100011753_100011912 | 0.53 |
Chrdl2 |
chordin-like 2 |
1918 |
0.27 |
| chr10_96652905_96653070 | 0.53 |
Btg1 |
BTG anti-proliferation factor 1 |
35433 |
0.15 |
| chr3_36532798_36533054 | 0.53 |
Gm11549 |
predicted gene 11549 |
542 |
0.65 |
| chr7_24599264_24599441 | 0.52 |
Phldb3 |
pleckstrin homology like domain, family B, member 3 |
11411 |
0.08 |
| chr18_31966444_31966632 | 0.52 |
Lims2 |
LIM and senescent cell antigen like domains 2 |
10205 |
0.13 |
| chr8_70832563_70832735 | 0.52 |
Arrdc2 |
arrestin domain containing 2 |
3938 |
0.09 |
| chr4_155234884_155235204 | 0.52 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
12452 |
0.15 |
| chr19_61226929_61227456 | 0.52 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
44 |
0.96 |
| chr4_134722177_134722332 | 0.51 |
Man1c1 |
mannosidase, alpha, class 1C, member 1 |
17964 |
0.18 |
| chr5_28082564_28082715 | 0.51 |
Insig1 |
insulin induced gene 1 |
7973 |
0.17 |
| chr13_24292555_24292723 | 0.51 |
Gm11340 |
predicted gene 11340 |
834 |
0.49 |
| chr4_116838867_116839391 | 0.51 |
Rpl36-ps8 |
ribosomal protein L36, pseudogene 8 |
1092 |
0.3 |
| chr5_38635788_38635969 | 0.51 |
Zfp518b |
zinc finger protein 518B |
48872 |
0.11 |
| chr2_84260786_84261039 | 0.51 |
Gm13711 |
predicted gene 13711 |
57399 |
0.12 |
| chr1_65156847_65157132 | 0.51 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
18575 |
0.11 |
| chr7_12979741_12980348 | 0.51 |
Zfp446 |
zinc finger protein 446 |
524 |
0.55 |
| chr8_127515714_127515885 | 0.50 |
Gm6921 |
predicted pseudogene 6921 |
45636 |
0.17 |
| chr7_100077379_100077530 | 0.50 |
Pold3 |
polymerase (DNA-directed), delta 3, accessory subunit |
12583 |
0.16 |
| chr16_3843876_3844135 | 0.50 |
Zfp174 |
zinc finger protein 174 |
3263 |
0.13 |
| chr5_102051269_102051420 | 0.50 |
Gm43787 |
predicted gene 43787 |
6725 |
0.19 |
| chr19_5717043_5717240 | 0.50 |
Ehbp1l1 |
EH domain binding protein 1-like 1 |
562 |
0.47 |
| chr9_46221443_46221600 | 0.49 |
Sik3 |
SIK family kinase 3 |
1950 |
0.18 |
| chr9_119145228_119145440 | 0.49 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
4729 |
0.13 |
| chr6_38531580_38531761 | 0.49 |
Fmc1 |
formation of mitochondrial complex V assembly factor 1 |
1832 |
0.27 |
| chr19_53225949_53226100 | 0.49 |
Add3 |
adducin 3 (gamma) |
27978 |
0.12 |
| chr1_36525855_36526032 | 0.49 |
Gm38033 |
predicted gene, 38033 |
2294 |
0.14 |
| chr4_123645434_123645623 | 0.49 |
Gm12926 |
predicted gene 12926 |
15233 |
0.13 |
| chr19_3912299_3912583 | 0.48 |
Ndufs8 |
NADH:ubiquinone oxidoreductase core subunit S8 |
47 |
0.93 |
| chr11_88860733_88860908 | 0.48 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
2897 |
0.21 |
| chr7_134663410_134663580 | 0.48 |
Gm9333 |
predicted gene 9333 |
3160 |
0.3 |
| chr3_88237842_88237993 | 0.48 |
Gm3764 |
predicted gene 3764 |
9134 |
0.08 |
| chr11_95450705_95450876 | 0.48 |
Gm11522 |
predicted gene 11522 |
11931 |
0.15 |
| chr6_119466953_119467108 | 0.48 |
Wnt5b |
wingless-type MMTV integration site family, member 5B |
181 |
0.95 |
| chr3_145561026_145561180 | 0.48 |
Znhit6 |
zinc finger, HIT type 6 |
15102 |
0.19 |
| chr4_139975184_139975335 | 0.48 |
Klhdc7a |
kelch domain containing 7A |
7233 |
0.17 |
| chr2_152737377_152737555 | 0.47 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
1215 |
0.31 |
| chr8_107186477_107186628 | 0.47 |
Cyb5b |
cytochrome b5 type B |
20637 |
0.15 |
| chr6_149144321_149144479 | 0.47 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
2754 |
0.18 |
| chr7_112188591_112188751 | 0.46 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
29614 |
0.19 |
| chr5_137358508_137358672 | 0.46 |
Ephb4 |
Eph receptor B4 |
8077 |
0.09 |
| chr8_25819139_25819316 | 0.46 |
Ash2l |
ASH2 like histone lysine methyltransferase complex subunit |
4348 |
0.13 |
| chr17_15050732_15050898 | 0.46 |
Ermard |
ER membrane associated RNA degradation |
470 |
0.74 |
| chr4_150732960_150733140 | 0.46 |
Gm16079 |
predicted gene 16079 |
54258 |
0.11 |
| chr16_33761869_33762022 | 0.46 |
Heg1 |
heart development protein with EGF-like domains 1 |
5422 |
0.2 |
| chr1_133084226_133084395 | 0.46 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
15731 |
0.13 |
| chr8_70128792_70128964 | 0.45 |
Nr2c2ap |
nuclear receptor 2C2-associated protein |
2455 |
0.12 |
| chr4_62778005_62778291 | 0.45 |
Gm24117 |
predicted gene, 24117 |
34280 |
0.14 |
| chr1_51761256_51761433 | 0.45 |
Myo1b |
myosin IB |
3126 |
0.26 |
| chr7_140119812_140119963 | 0.45 |
Echs1 |
enoyl Coenzyme A hydratase, short chain, 1, mitochondrial |
3411 |
0.11 |
| chr17_12389802_12390011 | 0.45 |
Plg |
plasminogen |
11247 |
0.16 |
| chr7_121907300_121907714 | 0.45 |
Scnn1b |
sodium channel, nonvoltage-gated 1 beta |
8279 |
0.18 |
| chr12_21171901_21172069 | 0.45 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
60031 |
0.09 |
| chr17_31116179_31116343 | 0.44 |
Abcg1 |
ATP binding cassette subfamily G member 1 |
5296 |
0.13 |
| chr9_86721274_86721452 | 0.44 |
Gm37484 |
predicted gene, 37484 |
9359 |
0.11 |
| chr17_15019435_15019586 | 0.44 |
Gm3435 |
predicted gene 3435 |
9175 |
0.12 |
| chr8_119817612_119817763 | 0.43 |
Cotl1 |
coactosin-like 1 (Dictyostelium) |
5030 |
0.19 |
| chr9_31251875_31252037 | 0.43 |
Gm7244 |
predicted gene 7244 |
22865 |
0.14 |
| chr5_113209325_113209655 | 0.43 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
11746 |
0.13 |
| chr16_4866490_4866641 | 0.43 |
4930562C15Rik |
RIKEN cDNA 4930562C15 gene |
2242 |
0.21 |
| chr2_75715885_75716036 | 0.43 |
E030042O20Rik |
RIKEN cDNA E030042O20 gene |
11190 |
0.12 |
| chr8_41373789_41374214 | 0.43 |
Asah1 |
N-acylsphingosine amidohydrolase 1 |
634 |
0.78 |
| chr8_68048016_68048207 | 0.42 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
14116 |
0.23 |
| chr3_85864106_85864273 | 0.42 |
Gm37240 |
predicted gene, 37240 |
23179 |
0.1 |
| chr17_34190794_34190960 | 0.42 |
Tap1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
125 |
0.88 |
| chr3_69664425_69664589 | 0.42 |
Gm37380 |
predicted gene, 37380 |
2741 |
0.24 |
| chr17_31940377_31940572 | 0.42 |
Gm17572 |
predicted gene, 17572 |
10060 |
0.14 |
| chr6_121048740_121049060 | 0.42 |
Gm4651 |
predicted gene 4651 |
15564 |
0.16 |
| chr11_32225384_32225575 | 0.42 |
Mpg |
N-methylpurine-DNA glycosylase |
1026 |
0.36 |
| chr16_90832098_90832268 | 0.42 |
Eva1c |
eva-1 homolog C (C. elegans) |
1070 |
0.43 |
| chr9_35107299_35107484 | 0.41 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
724 |
0.6 |
| chr9_110890046_110890620 | 0.41 |
Als2cl |
ALS2 C-terminal like |
2338 |
0.14 |
| chr14_76556939_76557219 | 0.41 |
Serp2 |
stress-associated endoplasmic reticulum protein family member 2 |
190 |
0.95 |
| chr15_39853276_39853629 | 0.41 |
Dpys |
dihydropyrimidinase |
4015 |
0.26 |
| chr14_8191471_8191629 | 0.41 |
Pdhb |
pyruvate dehydrogenase (lipoamide) beta |
18525 |
0.16 |
| chr5_36120892_36121056 | 0.41 |
Psapl1 |
prosaposin-like 1 |
83047 |
0.09 |
| chr5_72857104_72857283 | 0.41 |
Tec |
tec protein tyrosine kinase |
11203 |
0.16 |
| chr17_34822031_34822194 | 0.40 |
C4a |
complement component 4A (Rodgers blood group) |
1342 |
0.16 |
| chr12_103311534_103311702 | 0.40 |
Fam181a |
family with sequence similarity 181, member A |
643 |
0.58 |
| chr7_113214865_113215031 | 0.40 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
7391 |
0.23 |
| chr4_135787434_135787634 | 0.40 |
Myom3 |
myomesin family, member 3 |
10656 |
0.14 |
| chr7_140864034_140864241 | 0.40 |
Ric8a |
RIC8 guanine nucleotide exchange factor A |
2964 |
0.08 |
| chr4_144899236_144899574 | 0.40 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
6186 |
0.23 |
| chr5_92134368_92134534 | 0.39 |
Uso1 |
USO1 vesicle docking factor |
3487 |
0.15 |
| chr19_57353272_57353492 | 0.39 |
Fam160b1 |
family with sequence similarity 160, member B1 |
7298 |
0.16 |
| chr6_38848060_38848236 | 0.39 |
Hipk2 |
homeodomain interacting protein kinase 2 |
10837 |
0.21 |
| chr10_77061312_77061487 | 0.39 |
Col18a1 |
collagen, type XVIII, alpha 1 |
1402 |
0.35 |
| chr6_85820600_85820889 | 0.39 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
210 |
0.49 |
| chr4_135269728_135270123 | 0.38 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
2889 |
0.19 |
| chr7_30361407_30361626 | 0.38 |
Lrfn3 |
leucine rich repeat and fibronectin type III domain containing 3 |
1256 |
0.21 |
| chr7_44256470_44256632 | 0.38 |
Acp4 |
acid phosphatase 4 |
654 |
0.31 |
| chr7_81599363_81599874 | 0.38 |
Gm45698 |
predicted gene 45698 |
8724 |
0.11 |
| chr17_5029352_5029503 | 0.38 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
11151 |
0.25 |
| chr3_28695654_28696216 | 0.38 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
1968 |
0.31 |
| chr3_144714230_144714412 | 0.38 |
Sh3glb1 |
SH3-domain GRB2-like B1 (endophilin) |
5789 |
0.15 |
| chr1_62715731_62715926 | 0.37 |
Gm29083 |
predicted gene 29083 |
2245 |
0.29 |
| chr2_31504934_31505459 | 0.37 |
Ass1 |
argininosuccinate synthetase 1 |
4470 |
0.21 |
| chr10_79933831_79934226 | 0.37 |
Arid3a |
AT rich interactive domain 3A (BRIGHT-like) |
3604 |
0.07 |
| chr8_35383605_35384049 | 0.37 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
7167 |
0.17 |
| chr11_78972853_78973033 | 0.37 |
Lgals9 |
lectin, galactose binding, soluble 9 |
2613 |
0.27 |
| chr6_17650329_17650480 | 0.37 |
Capza2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
10202 |
0.18 |
| chr5_93049247_93049419 | 0.37 |
Sowahb |
sosondowah ankyrin repeat domain family member B |
4311 |
0.15 |
| chr8_122550942_122551215 | 0.37 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
251 |
0.84 |
| chr12_78209771_78209951 | 0.37 |
Gm6657 |
predicted gene 6657 |
8895 |
0.15 |
| chr7_98428680_98428844 | 0.37 |
Gm44980 |
predicted gene 44980 |
8757 |
0.12 |
| chr7_134192391_134192592 | 0.36 |
Adam12 |
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
19547 |
0.21 |
| chr6_48388505_48388930 | 0.36 |
Gm7880 |
predicted gene 7880 |
6657 |
0.12 |
| chr4_117303634_117303831 | 0.36 |
Rnf220 |
ring finger protein 220 |
3704 |
0.16 |
| chr6_117656074_117656246 | 0.36 |
Gm45083 |
predicted gene 45083 |
43590 |
0.13 |
| chr19_3341372_3341523 | 0.36 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
180 |
0.92 |
| chr6_54726603_54727071 | 0.36 |
Gm44008 |
predicted gene, 44008 |
33191 |
0.14 |
| chr14_31392223_31392522 | 0.36 |
Sh3bp5 |
SH3-domain binding protein 5 (BTK-associated) |
15013 |
0.13 |
| chr5_137318642_137318812 | 0.36 |
Slc12a9 |
solute carrier family 12 (potassium/chloride transporters), member 9 |
3785 |
0.09 |
| chr2_181147120_181147301 | 0.36 |
Eef1a2 |
eukaryotic translation elongation factor 1 alpha 2 |
9804 |
0.11 |
| chr6_23121749_23122234 | 0.36 |
Aass |
aminoadipate-semialdehyde synthase |
5589 |
0.18 |
| chr1_120136442_120136748 | 0.35 |
3110009E18Rik |
RIKEN cDNA 3110009E18 gene |
14004 |
0.18 |
| chr9_62380562_62380925 | 0.35 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
7304 |
0.22 |
| chr12_82121365_82121534 | 0.35 |
n-R5s62 |
nuclear encoded rRNA 5S 62 |
29019 |
0.17 |
| chr11_117278201_117278394 | 0.35 |
Septin9 |
septin 9 |
12051 |
0.18 |
| chr11_117321409_117321564 | 0.35 |
Septin9 |
septin 9 |
4947 |
0.22 |
| chr15_82747379_82747542 | 0.35 |
Gm49433 |
predicted gene, 49433 |
5344 |
0.1 |
| chr2_103599180_103599608 | 0.34 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
33084 |
0.16 |
| chr9_107569247_107569427 | 0.34 |
Hyal2 |
hyaluronoglucosaminidase 2 |
58 |
0.87 |
| chr19_7057314_7057476 | 0.34 |
Macrod1 |
mono-ADP ribosylhydrolase 1 |
585 |
0.58 |
| chr15_31336506_31336683 | 0.34 |
Ankrd33b |
ankyrin repeat domain 33B |
11345 |
0.16 |
| chr10_62966160_62966539 | 0.34 |
Dna2 |
DNA replication helicase/nuclease 2 |
373 |
0.75 |
| chr2_164131659_164131883 | 0.34 |
Gm11455 |
predicted gene 11455 |
3932 |
0.13 |
| chr17_45801293_45801458 | 0.34 |
Gm35692 |
predicted gene, 35692 |
20948 |
0.14 |
| chr17_80167519_80167694 | 0.34 |
Galm |
galactose mutarotase |
22454 |
0.13 |
| chr18_35256811_35256978 | 0.34 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
4573 |
0.19 |
| chr9_109959682_109959984 | 0.34 |
Map4 |
microtubule-associated protein 4 |
9184 |
0.12 |
| chr3_104623903_104624054 | 0.34 |
Gm43582 |
predicted gene 43582 |
10679 |
0.1 |
| chr7_125578941_125579144 | 0.34 |
Gm44876 |
predicted gene 44876 |
6925 |
0.16 |
| chr14_34663553_34663710 | 0.33 |
Gm49024 |
predicted gene, 49024 |
1924 |
0.23 |
| chr10_62920860_62921219 | 0.33 |
Slc25a16 |
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
359 |
0.75 |
| chr11_83850528_83850683 | 0.33 |
Hnf1b |
HNF1 homeobox B |
250 |
0.85 |
| chr6_33003982_33004151 | 0.33 |
Gm14540 |
predicted gene 14540 |
34402 |
0.16 |
| chr15_82393638_82393839 | 0.33 |
Mir7684 |
microRNA 7684 |
240 |
0.49 |
| chr8_121645240_121645446 | 0.33 |
Zcchc14 |
zinc finger, CCHC domain containing 14 |
7558 |
0.12 |
| chr17_45588490_45588676 | 0.33 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
158 |
0.89 |
| chr9_23305796_23305965 | 0.33 |
Bmper |
BMP-binding endothelial regulator |
68052 |
0.14 |
| chr6_38230014_38230184 | 0.33 |
D630045J12Rik |
RIKEN cDNA D630045J12 gene |
23856 |
0.16 |
| chr19_57537517_57537807 | 0.33 |
Gm50285 |
predicted gene, 50285 |
6561 |
0.15 |
| chr17_31852879_31853048 | 0.33 |
Sik1 |
salt inducible kinase 1 |
1388 |
0.38 |
| chr2_113610286_113610454 | 0.33 |
Fmn1 |
formin 1 |
74720 |
0.09 |
| chr19_16585537_16585696 | 0.33 |
Gm50363 |
predicted gene, 50363 |
13148 |
0.22 |
| chr6_48038853_48039010 | 0.33 |
Zfp777 |
zinc finger protein 777 |
5312 |
0.14 |
| chr2_32775126_32775277 | 0.33 |
Ttc16 |
tetratricopeptide repeat domain 16 |
129 |
0.82 |
| chr14_28656813_28657238 | 0.33 |
Gm35164 |
predicted gene, 35164 |
7266 |
0.27 |
| chr2_27462999_27463172 | 0.33 |
Brd3 |
bromodomain containing 3 |
1031 |
0.44 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.3 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.2 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.1 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.2 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.0 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.2 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:2000391 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) |
| 0.0 | 0.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0006522 | alanine metabolic process(GO:0006522) pyruvate family amino acid metabolic process(GO:0009078) L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.0 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.0 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 | 0.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.3 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.0 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.0 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0008334 | histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.0 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.0 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0043733 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0018647 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.4 | GO:0018731 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.0 | GO:0051723 | protein methylesterase activity(GO:0051723) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.0 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.0 | 0.1 | GO:0042300 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |