Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
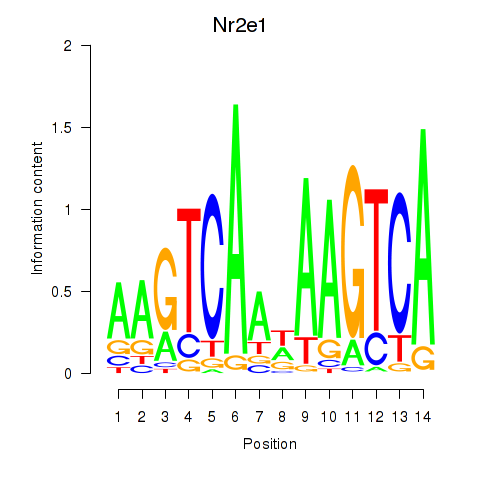
Results for Nr2e1
Z-value: 0.89
Transcription factors associated with Nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e1
|
ENSMUSG00000019803.5 | nuclear receptor subfamily 2, group E, member 1 |
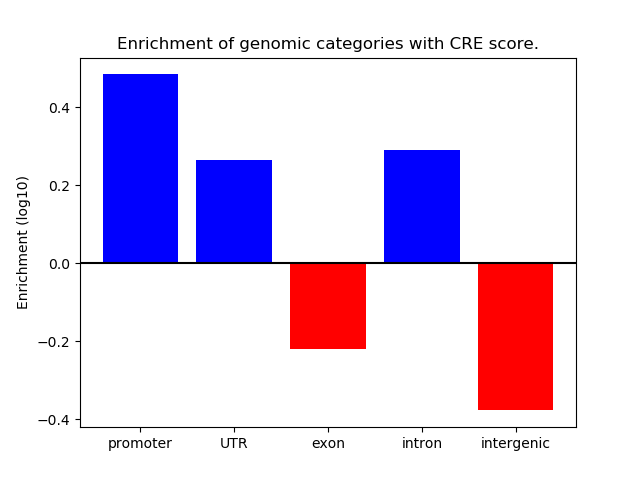
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_42538714_42538885 | Nr2e1 | 30029 | 0.145705 | 0.80 | 5.4e-02 | Click! |
Activity of the Nr2e1 motif across conditions
Conditions sorted by the z-value of the Nr2e1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
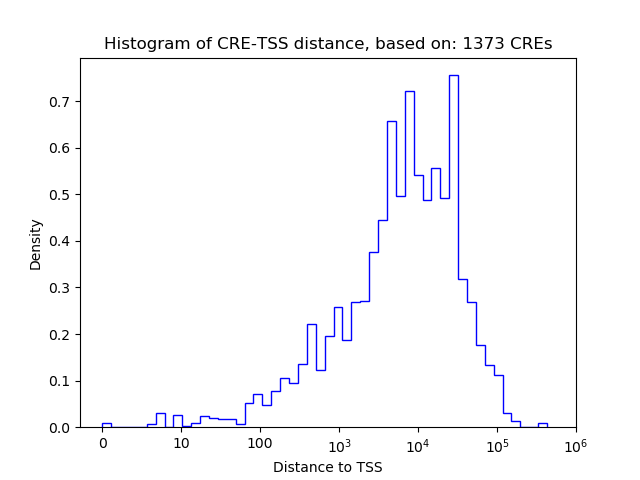
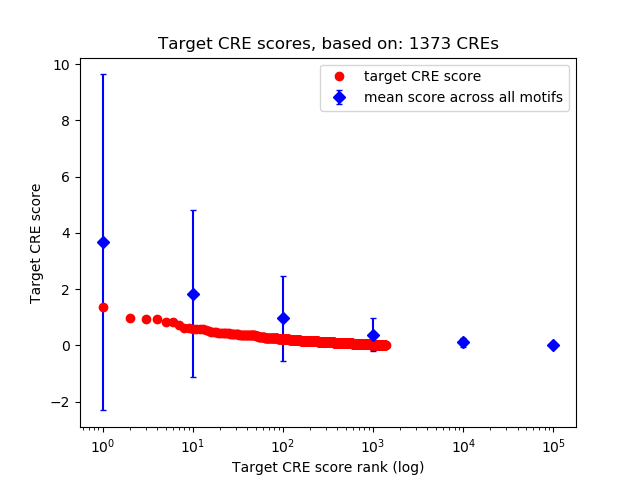
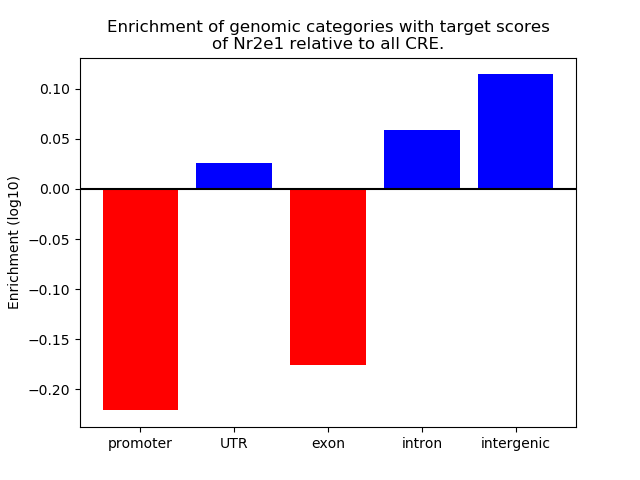
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_146107597_146107748 | 1.37 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
28405 |
0.11 |
| chr5_118292276_118292427 | 0.98 |
Gm25076 |
predicted gene, 25076 |
25902 |
0.16 |
| chr5_145982849_145983558 | 0.94 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
8440 |
0.13 |
| chr3_88628445_88628599 | 0.93 |
Arhgef2 |
rho/rac guanine nucleotide exchange factor (GEF) 2 |
920 |
0.37 |
| chr10_115348229_115348396 | 0.83 |
Tmem19 |
transmembrane protein 19 |
752 |
0.58 |
| chr10_122408778_122408952 | 0.82 |
Gm36041 |
predicted gene, 36041 |
21973 |
0.18 |
| chr9_78022696_78022864 | 0.74 |
Gm47829 |
predicted gene, 47829 |
5165 |
0.12 |
| chr14_19839858_19840045 | 0.61 |
D14Ertd670e |
DNA segment, Chr 14, ERATO Doi 670, expressed |
1026 |
0.48 |
| chr8_73114845_73115014 | 0.60 |
Large1 |
LARGE xylosyl- and glucuronyltransferase 1 |
16016 |
0.24 |
| chr10_28078586_28078966 | 0.59 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
3766 |
0.28 |
| chr4_118129721_118129877 | 0.58 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
5083 |
0.18 |
| chr11_107109633_107109941 | 0.58 |
Bptf |
bromodomain PHD finger transcription factor |
22135 |
0.14 |
| chr15_8968437_8968609 | 0.56 |
Ranbp3l |
RAN binding protein 3-like |
97 |
0.97 |
| chr5_98996002_98996174 | 0.54 |
Gm3470 |
predicted gene 3470 |
26134 |
0.2 |
| chr3_101953485_101953642 | 0.49 |
Slc22a15 |
solute carrier family 22 (organic anion/cation transporter), member 15 |
29110 |
0.17 |
| chr17_24797110_24797261 | 0.48 |
Meiob |
meiosis specific with OB domains |
7104 |
0.09 |
| chr7_101491003_101491309 | 0.46 |
Pde2a |
phosphodiesterase 2A, cGMP-stimulated |
9871 |
0.14 |
| chr19_41838995_41839153 | 0.46 |
Frat2 |
frequently rearranged in advanced T cell lymphomas 2 |
9058 |
0.14 |
| chr12_84939464_84939615 | 0.45 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
3839 |
0.15 |
| chr9_66327050_66327224 | 0.45 |
Herc1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
23313 |
0.17 |
| chr11_50733265_50733435 | 0.45 |
Gm12200 |
predicted gene 12200 |
7806 |
0.16 |
| chr6_30286706_30286895 | 0.44 |
Ube2h |
ubiquitin-conjugating enzyme E2H |
17722 |
0.14 |
| chr19_33094946_33095121 | 0.44 |
Gm29863 |
predicted gene, 29863 |
24961 |
0.18 |
| chr14_60636670_60636838 | 0.43 |
Spata13 |
spermatogenesis associated 13 |
1999 |
0.38 |
| chr5_62037605_62037780 | 0.43 |
Gm42430 |
predicted gene 42430 |
33472 |
0.23 |
| chr15_90400394_90400702 | 0.41 |
Gm36480 |
predicted gene, 36480 |
46842 |
0.18 |
| chr15_54923012_54923175 | 0.41 |
Enpp2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
480 |
0.81 |
| chr11_83271635_83271942 | 0.41 |
Gm23444 |
predicted gene, 23444 |
290 |
0.8 |
| chr8_94217878_94218059 | 0.40 |
Nup93 |
nucleoporin 93 |
3362 |
0.14 |
| chr2_67957572_67957902 | 0.40 |
Gm37964 |
predicted gene, 37964 |
59141 |
0.14 |
| chr11_70748249_70748424 | 0.39 |
Gm12320 |
predicted gene 12320 |
6815 |
0.09 |
| chr14_74733504_74733933 | 0.39 |
Esd |
esterase D/formylglutathione hydrolase |
1334 |
0.47 |
| chr2_119318398_119318585 | 0.38 |
Gm14207 |
predicted gene 14207 |
7052 |
0.12 |
| chr6_73513110_73513275 | 0.38 |
Gm4374 |
predicted gene 4374 |
16046 |
0.19 |
| chr7_84135967_84136134 | 0.38 |
Abhd17c |
abhydrolase domain containing 17C |
11765 |
0.15 |
| chr9_80777245_80777426 | 0.38 |
Gm27224 |
predicted gene 27224 |
25316 |
0.22 |
| chr10_34136560_34136736 | 0.37 |
Calhm6 |
calcium homeostasis modulator family member 6 |
8664 |
0.13 |
| chr9_110573946_110574097 | 0.37 |
Setd2 |
SET domain containing 2 |
137 |
0.94 |
| chr16_24223745_24224269 | 0.37 |
Gm31814 |
predicted gene, 31814 |
7483 |
0.23 |
| chr7_49765686_49765845 | 0.37 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
6612 |
0.22 |
| chr11_109734187_109734610 | 0.37 |
Fam20a |
family with sequence similarity 20, member A |
12119 |
0.17 |
| chr1_95590205_95590368 | 0.36 |
Gm37642 |
predicted gene, 37642 |
58222 |
0.13 |
| chr10_84055512_84055676 | 0.36 |
Gm37908 |
predicted gene, 37908 |
6164 |
0.21 |
| chr9_53315738_53315899 | 0.36 |
Exph5 |
exophilin 5 |
14148 |
0.18 |
| chr8_117720839_117721024 | 0.36 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
5 |
0.97 |
| chr6_92922025_92922180 | 0.36 |
9530026P05Rik |
RIKEN cDNA 9530026P05 gene |
18227 |
0.19 |
| chr7_98537643_98537832 | 0.36 |
A630091E08Rik |
RIKEN cDNA A630091E08 gene |
25253 |
0.12 |
| chr7_126230828_126230994 | 0.35 |
Gm44703 |
predicted gene 44703 |
11716 |
0.12 |
| chr11_87735822_87735981 | 0.35 |
Supt4a |
SPT4A, DSIF elongation factor subunit |
1651 |
0.19 |
| chr16_93325022_93325192 | 0.34 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
28082 |
0.17 |
| chr10_61137486_61137750 | 0.33 |
Sgpl1 |
sphingosine phosphate lyase 1 |
9280 |
0.15 |
| chr16_33837321_33837485 | 0.32 |
Itgb5 |
integrin beta 5 |
7726 |
0.17 |
| chr12_103820890_103821429 | 0.32 |
Gm8893 |
predicted gene 8893 |
1084 |
0.34 |
| chr16_96943868_96944022 | 0.31 |
Gm32432 |
predicted gene, 32432 |
25188 |
0.26 |
| chr13_37638048_37638218 | 0.30 |
Gm40916 |
predicted gene, 40916 |
10103 |
0.12 |
| chr15_82788288_82788464 | 0.30 |
Cyp2d26 |
cytochrome P450, family 2, subfamily d, polypeptide 26 |
5846 |
0.1 |
| chr6_31453844_31454029 | 0.29 |
Mkln1 |
muskelin 1, intracellular mediator containing kelch motifs |
14184 |
0.19 |
| chr8_122318051_122318212 | 0.29 |
Zfpm1 |
zinc finger protein, multitype 1 |
10811 |
0.13 |
| chr2_3579492_3579678 | 0.29 |
Fam107b |
family with sequence similarity 107, member B |
9097 |
0.15 |
| chr11_29532358_29532531 | 0.28 |
Mtif2 |
mitochondrial translational initiation factor 2 |
846 |
0.45 |
| chr13_45262109_45262392 | 0.28 |
Gm34466 |
predicted gene, 34466 |
5461 |
0.23 |
| chr18_4827364_4827665 | 0.28 |
Gm10556 |
predicted gene 10556 |
15028 |
0.25 |
| chr2_22587937_22588311 | 0.27 |
Gm13341 |
predicted gene 13341 |
162 |
0.91 |
| chr7_123125696_123126050 | 0.27 |
Tnrc6a |
trinucleotide repeat containing 6a |
1617 |
0.43 |
| chr3_121904528_121904954 | 0.27 |
Gm42593 |
predicted gene 42593 |
41899 |
0.12 |
| chr1_88386138_88386312 | 0.27 |
Spp2 |
secreted phosphoprotein 2 |
20736 |
0.14 |
| chr3_155090080_155090231 | 0.27 |
Fpgt |
fucose-1-phosphate guanylyltransferase |
364 |
0.84 |
| chr11_53418376_53418584 | 0.27 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
4690 |
0.09 |
| chr18_60748821_60749236 | 0.26 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
378 |
0.77 |
| chr1_72809182_72809347 | 0.26 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
15239 |
0.21 |
| chrX_168959751_168959918 | 0.26 |
Arhgap6 |
Rho GTPase activating protein 6 |
76777 |
0.11 |
| chr15_46625156_46625324 | 0.26 |
4930548G14Rik |
RIKEN cDNA 4930548G14 gene |
216 |
0.97 |
| chr14_56746132_56746283 | 0.26 |
Pspc1 |
paraspeckle protein 1 |
15660 |
0.12 |
| chr15_82668190_82668753 | 0.25 |
Cyp2d35-ps |
cytochrome P450, family 2, subfamily d, member 35, pseudogene |
13760 |
0.07 |
| chr15_11394872_11395043 | 0.25 |
Tars |
threonyl-tRNA synthetase |
2687 |
0.35 |
| chr16_32728387_32728559 | 0.25 |
Gm9556 |
predicted gene 9556 |
1807 |
0.27 |
| chr16_29988549_29988737 | 0.25 |
Gm1968 |
predicted gene 1968 |
9459 |
0.16 |
| chr6_128538055_128538238 | 0.25 |
A2ml1 |
alpha-2-macroglobulin like 1 |
5348 |
0.1 |
| chr14_25751185_25751350 | 0.25 |
Zcchc24 |
zinc finger, CCHC domain containing 24 |
17772 |
0.14 |
| chr17_48959982_48960147 | 0.25 |
Lrfn2 |
leucine rich repeat and fibronectin type III domain containing 2 |
27685 |
0.23 |
| chr16_24370710_24370861 | 0.25 |
Gm41434 |
predicted gene, 41434 |
553 |
0.74 |
| chr7_80340519_80340853 | 0.25 |
Unc45a |
unc-45 myosin chaperone A |
467 |
0.64 |
| chr12_3912062_3912221 | 0.25 |
Dnmt3a |
DNA methyltransferase 3A |
5325 |
0.17 |
| chr6_119139630_119139800 | 0.24 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
31142 |
0.19 |
| chr5_130044523_130044702 | 0.24 |
Crcp |
calcitonin gene-related peptide-receptor component protein |
2408 |
0.2 |
| chr9_48618198_48618386 | 0.24 |
Nnmt |
nicotinamide N-methyltransferase |
13139 |
0.24 |
| chr2_156175837_156176038 | 0.24 |
Rbm39 |
RNA binding motif protein 39 |
1446 |
0.28 |
| chr4_26470101_26470279 | 0.24 |
Gm11903 |
predicted gene 11903 |
27729 |
0.19 |
| chr15_25069819_25069985 | 0.24 |
Gm2824 |
predicted gene 2824 |
42544 |
0.15 |
| chrX_101953397_101953559 | 0.24 |
Nhsl2 |
NHS-like 2 |
8319 |
0.18 |
| chr19_5810333_5810765 | 0.24 |
Malat1 |
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
7877 |
0.08 |
| chr16_21474778_21474944 | 0.24 |
Vps8 |
VPS8 CORVET complex subunit |
4674 |
0.3 |
| chr19_41302182_41302348 | 0.23 |
Tm9sf3 |
transmembrane 9 superfamily member 3 |
38268 |
0.17 |
| chr10_110707462_110707627 | 0.23 |
E2f7 |
E2F transcription factor 7 |
37895 |
0.16 |
| chr4_141513828_141514048 | 0.23 |
Spen |
spen family transcription repressor |
2971 |
0.19 |
| chr18_74624820_74624979 | 0.23 |
Myo5b |
myosin VB |
8278 |
0.25 |
| chr11_102742268_102742437 | 0.23 |
Gm16342 |
predicted gene 16342 |
9947 |
0.11 |
| chr11_22443828_22443986 | 0.23 |
Gm12050 |
predicted gene 12050 |
4132 |
0.24 |
| chr15_59373722_59374046 | 0.23 |
Washc5 |
WASH complex subunit 5 |
283 |
0.57 |
| chr14_51063014_51063572 | 0.23 |
Rnase12 |
ribonuclease, RNase A family, 12 (non-active) |
5632 |
0.09 |
| chr18_11858447_11858616 | 0.22 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
14231 |
0.16 |
| chr3_65660431_65660599 | 0.22 |
Mir8120 |
microRNA 8120 |
1227 |
0.38 |
| chr15_63364555_63364706 | 0.22 |
Gm41335 |
predicted gene, 41335 |
7611 |
0.27 |
| chr15_89477372_89477550 | 0.22 |
Arsa |
arylsulfatase A |
36 |
0.77 |
| chr13_58058115_58058280 | 0.22 |
Mir874 |
microRNA 874 |
34997 |
0.13 |
| chr14_20830071_20830269 | 0.22 |
Plau |
plasminogen activator, urokinase |
6490 |
0.15 |
| chr16_22683678_22683832 | 0.22 |
Gm8118 |
predicted gene 8118 |
2439 |
0.27 |
| chr10_127937077_127937501 | 0.21 |
Gm15900 |
predicted gene 15900 |
11271 |
0.09 |
| chr13_69583732_69583897 | 0.21 |
Srd5a1 |
steroid 5 alpha-reductase 1 |
11015 |
0.12 |
| chr17_51852545_51852696 | 0.21 |
Gm37266 |
predicted gene, 37266 |
2381 |
0.17 |
| chr1_160730069_160730244 | 0.21 |
Gm38329 |
predicted gene, 38329 |
1996 |
0.22 |
| chr3_97631636_97631799 | 0.21 |
Fmo5 |
flavin containing monooxygenase 5 |
2834 |
0.19 |
| chr18_46630413_46630795 | 0.21 |
Gm3734 |
predicted gene 3734 |
203 |
0.92 |
| chr4_61777961_61778274 | 0.21 |
Mup19 |
major urinary protein 19 |
4107 |
0.15 |
| chr8_103468201_103468434 | 0.21 |
Gm45277 |
predicted gene 45277 |
95306 |
0.08 |
| chr1_15118618_15118800 | 0.21 |
Gm26345 |
predicted gene, 26345 |
99550 |
0.08 |
| chr9_45030162_45030366 | 0.21 |
Mpzl2 |
myelin protein zero-like 2 |
9535 |
0.08 |
| chr5_70735708_70735881 | 0.20 |
Gabrg1 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
106410 |
0.07 |
| chr8_33885103_33885396 | 0.20 |
Gm26978 |
predicted gene, 26978 |
498 |
0.78 |
| chr2_48539799_48539950 | 0.20 |
Gm13481 |
predicted gene 13481 |
82629 |
0.1 |
| chr3_95653729_95653883 | 0.20 |
Mcl1 |
myeloid cell leukemia sequence 1 |
4982 |
0.11 |
| chr9_116947417_116947569 | 0.20 |
Gm18489 |
predicted gene, 18489 |
111665 |
0.07 |
| chr15_66074244_66074502 | 0.20 |
Gm27153 |
predicted gene 27153 |
179 |
0.96 |
| chr4_3829069_3829220 | 0.20 |
Gm24016 |
predicted gene, 24016 |
6002 |
0.13 |
| chr4_32607361_32607706 | 0.20 |
Gm11933 |
predicted gene 11933 |
3818 |
0.16 |
| chr11_85618066_85618217 | 0.20 |
Bcas3 |
breast carcinoma amplified sequence 3 |
22563 |
0.21 |
| chr5_87087250_87087541 | 0.20 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
3762 |
0.14 |
| chr10_94080899_94081055 | 0.20 |
Fgd6 |
FYVE, RhoGEF and PH domain containing 6 |
8435 |
0.13 |
| chr8_112018421_112018578 | 0.20 |
Gm6793 |
predicted gene 6793 |
3491 |
0.17 |
| chr15_62067048_62067348 | 0.20 |
Pvt1 |
Pvt1 oncogene |
27941 |
0.22 |
| chr2_168955956_168956111 | 0.19 |
Zfp64 |
zinc finger protein 64 |
446 |
0.81 |
| chr2_115093304_115093458 | 0.19 |
Gm28493 |
predicted gene 28493 |
42169 |
0.17 |
| chr7_19797036_19797215 | 0.19 |
Cblc |
Casitas B-lineage lymphoma c |
316 |
0.74 |
| chr11_112720588_112720957 | 0.19 |
BC006965 |
cDNA sequence BC006965 |
9409 |
0.26 |
| chr4_104530164_104530485 | 0.19 |
Dab1 |
disabled 1 |
18251 |
0.29 |
| chr4_60497810_60497972 | 0.19 |
Mup1 |
major urinary protein 1 |
1441 |
0.3 |
| chr5_139338482_139338648 | 0.19 |
Cox19 |
cytochrome c oxidase assembly protein 19 |
1010 |
0.41 |
| chr17_13802909_13803139 | 0.19 |
Tcte2 |
t-complex-associated testis expressed 2 |
41199 |
0.11 |
| chr11_117242878_117243030 | 0.18 |
Septin9 |
septin 9 |
10669 |
0.18 |
| chr9_21799040_21799201 | 0.18 |
Kank2 |
KN motif and ankyrin repeat domains 2 |
376 |
0.76 |
| chr11_62568400_62568605 | 0.18 |
Gm12280 |
predicted gene 12280 |
2119 |
0.14 |
| chr1_24615703_24615868 | 0.18 |
Gm28661 |
predicted gene 28661 |
220 |
0.62 |
| chr7_140748467_140748790 | 0.18 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
15111 |
0.09 |
| chr8_119824884_119825204 | 0.18 |
Cotl1 |
coactosin-like 1 (Dictyostelium) |
2327 |
0.27 |
| chr2_169634592_169634925 | 0.18 |
Tshz2 |
teashirt zinc finger family member 2 |
1082 |
0.55 |
| chr4_60135275_60135767 | 0.18 |
Mup2 |
major urinary protein 2 |
4336 |
0.2 |
| chr16_77329783_77330099 | 0.18 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
437 |
0.81 |
| chr11_83743719_83744166 | 0.18 |
Wfdc21 |
WAP four-disulfide core domain 21 |
2998 |
0.13 |
| chr9_54738475_54738626 | 0.17 |
Wdr61 |
WD repeat domain 61 |
4031 |
0.17 |
| chr3_125754703_125754854 | 0.17 |
Ndst4 |
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
44692 |
0.2 |
| chr12_86324285_86324473 | 0.17 |
4732487G21Rik |
RIKEN cDNA 4732487G21 gene |
18373 |
0.17 |
| chr3_133765600_133766533 | 0.17 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr1_167381535_167381702 | 0.17 |
Mgst3 |
microsomal glutathione S-transferase 3 |
12223 |
0.14 |
| chr19_23142829_23142995 | 0.17 |
Klf9 |
Kruppel-like factor 9 |
732 |
0.62 |
| chr17_43418663_43418829 | 0.17 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
8266 |
0.22 |
| chr10_78266639_78266790 | 0.17 |
Gm47976 |
predicted gene, 47976 |
639 |
0.52 |
| chr15_10901196_10901355 | 0.17 |
Gm29742 |
predicted gene, 29742 |
16162 |
0.17 |
| chr1_86630817_86630968 | 0.17 |
Gm22955 |
predicted gene, 22955 |
6716 |
0.13 |
| chr17_88050390_88050623 | 0.17 |
Fbxo11 |
F-box protein 11 |
14381 |
0.19 |
| chr8_126615919_126616193 | 0.17 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
22070 |
0.21 |
| chr8_115719843_115720129 | 0.17 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
12192 |
0.23 |
| chr10_83244234_83244393 | 0.17 |
Slc41a2 |
solute carrier family 41, member 2 |
60062 |
0.11 |
| chr1_179836517_179836707 | 0.17 |
Ahctf1 |
AT hook containing transcription factor 1 |
32932 |
0.16 |
| chr16_23337603_23337782 | 0.17 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
47222 |
0.11 |
| chr16_93808092_93808456 | 0.17 |
Dop1b |
DOP1 leucine zipper like protein B |
1160 |
0.4 |
| chr2_135693224_135693399 | 0.17 |
Gm14211 |
predicted gene 14211 |
157 |
0.96 |
| chr8_10939998_10940171 | 0.17 |
Gm44955 |
predicted gene 44955 |
7806 |
0.11 |
| chr2_75563293_75563456 | 0.17 |
Gm13655 |
predicted gene 13655 |
70008 |
0.08 |
| chr14_73253316_73253480 | 0.16 |
Rb1 |
RB transcriptional corepressor 1 |
10923 |
0.17 |
| chr9_77927047_77927209 | 0.16 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
5196 |
0.17 |
| chr2_91794631_91794782 | 0.16 |
Ambra1 |
autophagy/beclin 1 regulator 1 |
16962 |
0.17 |
| chr11_120624046_120624203 | 0.16 |
Mir6936 |
microRNA 6936 |
752 |
0.26 |
| chr18_35704126_35704282 | 0.16 |
1700066B19Rik |
RIKEN cDNA 1700066B19 gene |
979 |
0.28 |
| chr7_81230207_81230362 | 0.16 |
Gm7180 |
predicted pseudogene 7180 |
7410 |
0.16 |
| chr4_60737134_60737406 | 0.16 |
Mup12 |
major urinary protein 12 |
4056 |
0.21 |
| chr3_104541230_104541625 | 0.16 |
Lrig2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
29509 |
0.1 |
| chr10_99555138_99555310 | 0.16 |
Gm48085 |
predicted gene, 48085 |
17155 |
0.18 |
| chr13_10445800_10445958 | 0.16 |
Gm47407 |
predicted gene, 47407 |
71808 |
0.1 |
| chr8_36712891_36713250 | 0.16 |
Dlc1 |
deleted in liver cancer 1 |
19984 |
0.26 |
| chr10_63373741_63373935 | 0.16 |
Sirt1 |
sirtuin 1 |
7866 |
0.13 |
| chr7_49475399_49475550 | 0.16 |
Gm38059 |
predicted gene, 38059 |
5762 |
0.25 |
| chr11_88059290_88059457 | 0.16 |
Gm15892 |
predicted gene 15892 |
307 |
0.84 |
| chr3_85821987_85822152 | 0.16 |
Fam160a1 |
family with sequence similarity 160, member A1 |
4778 |
0.2 |
| chr4_62087229_62087597 | 0.16 |
Mup3 |
major urinary protein 3 |
71 |
0.93 |
| chr5_16316197_16316406 | 0.16 |
Cacna2d1 |
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
9684 |
0.24 |
| chr6_72212736_72212914 | 0.16 |
Atoh8 |
atonal bHLH transcription factor 8 |
21712 |
0.14 |
| chr11_102543825_102543998 | 0.16 |
Mdk-ps1 |
midkine pseudogene 1 |
1791 |
0.21 |
| chr3_146581553_146581772 | 0.16 |
Uox |
urate oxidase |
11236 |
0.13 |
| chr9_66285727_66286140 | 0.16 |
Dapk2 |
death-associated protein kinase 2 |
17559 |
0.2 |
| chr6_99206656_99206831 | 0.16 |
Foxp1 |
forkhead box P1 |
41593 |
0.2 |
| chr1_89516107_89516298 | 0.16 |
Gm37297 |
predicted gene, 37297 |
32764 |
0.15 |
| chr8_128387567_128387718 | 0.16 |
Nrp1 |
neuropilin 1 |
28245 |
0.19 |
| chr5_102481207_102481593 | 0.16 |
Arhgap24 |
Rho GTPase activating protein 24 |
9 |
0.97 |
| chr17_28279592_28279743 | 0.15 |
Ppard |
peroxisome proliferator activator receptor delta |
7548 |
0.11 |
| chr4_60066252_60066431 | 0.15 |
Mup7 |
major urinary protein 7 |
4070 |
0.21 |
| chr2_174672606_174672877 | 0.15 |
Zfp831 |
zinc finger protein 831 |
29207 |
0.18 |
| chr9_20803282_20803448 | 0.15 |
Col5a3 |
collagen, type V, alpha 3 |
11702 |
0.11 |
| chr7_80024172_80024601 | 0.15 |
Zfp710 |
zinc finger protein 710 |
428 |
0.76 |
| chr5_145191503_145191838 | 0.15 |
Atp5j2 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
78 |
0.94 |
| chr11_113610401_113610567 | 0.15 |
Sstr2 |
somatostatin receptor 2 |
8858 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.3 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.0 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0014745 | negative regulation of muscle adaptation(GO:0014745) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0098911 | regulation of ventricular cardiac muscle cell action potential(GO:0098911) |
| 0.0 | 0.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.0 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0061626 | pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0043338 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |