Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
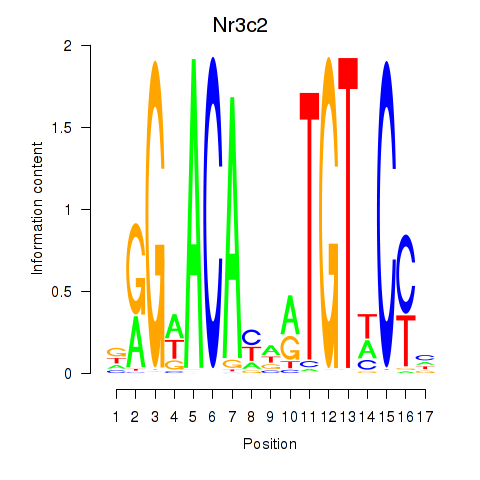
Results for Nr3c2
Z-value: 6.34
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSMUSG00000031618.7 | nuclear receptor subfamily 3, group C, member 2 |
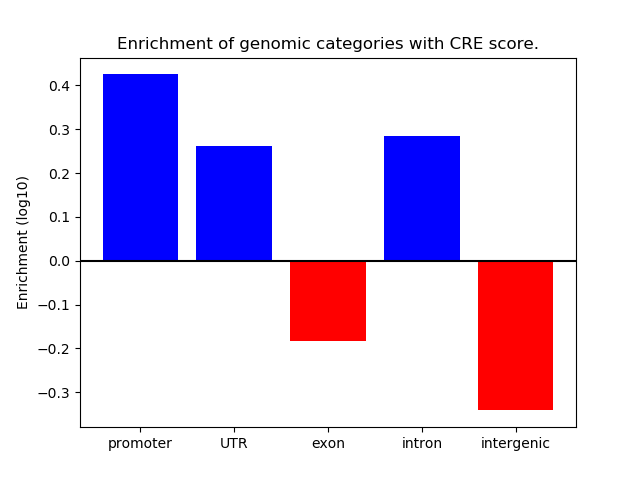
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
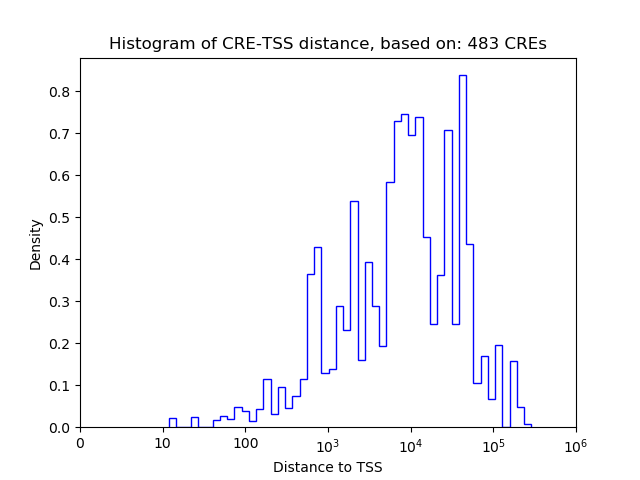
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_76935013_76935165 | Nr3c2 | 26817 | 0.191990 | 0.95 | 3.6e-03 | Click! |
| chr8_76902033_76902222 | Nr3c2 | 161 | 0.941199 | -0.94 | 4.9e-03 | Click! |
| chr8_77057050_77057484 | Nr3c2 | 70746 | 0.106913 | 0.94 | 5.7e-03 | Click! |
| chr8_76902272_76902430 | Nr3c2 | 4 | 0.973850 | -0.91 | 1.2e-02 | Click! |
| chr8_76983976_76984150 | Nr3c2 | 75791 | 0.098961 | 0.91 | 1.2e-02 | Click! |
Activity of the Nr3c2 motif across conditions
Conditions sorted by the z-value of the Nr3c2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
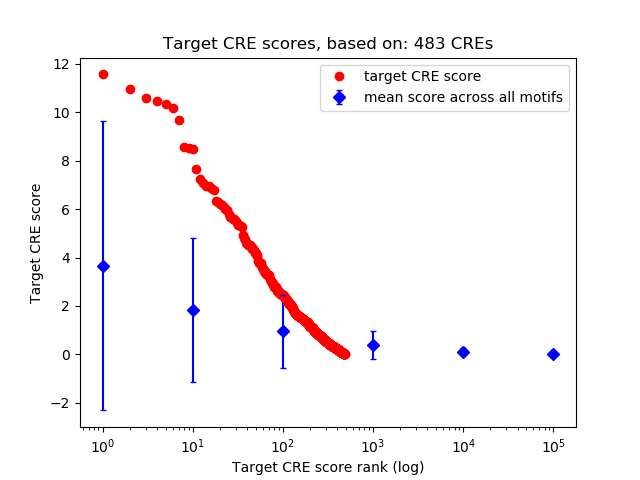
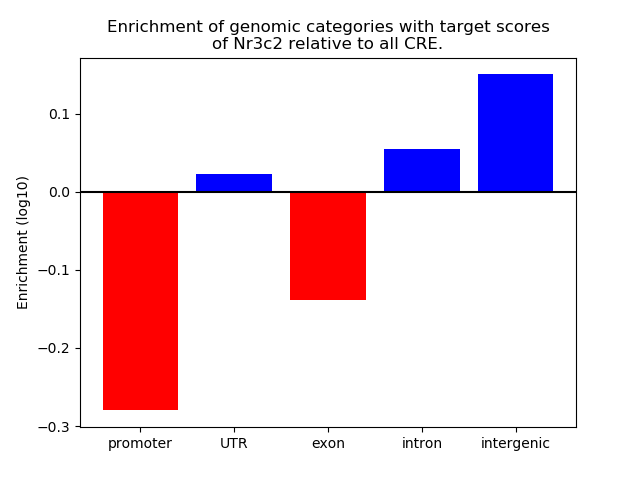
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_25884401_25884651 | 11.57 |
Gm6434 |
predicted gene 6434 |
2137 |
0.18 |
| chr1_162891903_162892071 | 10.98 |
Fmo2 |
flavin containing monooxygenase 2 |
5472 |
0.19 |
| chr18_54250751_54250951 | 10.58 |
Redrum |
Redrum, erythroid developmental long intergenic non-protein coding transcript |
171444 |
0.03 |
| chr2_160805166_160805505 | 10.45 |
Gm11447 |
predicted gene 11447 |
40278 |
0.11 |
| chr1_174765547_174765725 | 10.33 |
Fmn2 |
formin 2 |
51310 |
0.17 |
| chr6_149359539_149359690 | 10.18 |
Gm15784 |
predicted gene 15784 |
2072 |
0.23 |
| chr13_82200839_82201017 | 9.67 |
Gm48155 |
predicted gene, 48155 |
111171 |
0.07 |
| chr18_84054624_84054776 | 8.55 |
Tshz1 |
teashirt zinc finger family member 1 |
30375 |
0.14 |
| chr19_12708659_12708815 | 8.55 |
Gm15962 |
predicted gene 15962 |
12466 |
0.09 |
| chr12_83544904_83545068 | 8.50 |
Zfyve1 |
zinc finger, FYVE domain containing 1 |
6719 |
0.16 |
| chr1_55509753_55509917 | 7.68 |
Gm37382 |
predicted gene, 37382 |
49625 |
0.12 |
| chr7_126932811_126932965 | 7.23 |
Kctd13 |
potassium channel tetramerisation domain containing 13 |
2967 |
0.08 |
| chr17_28420256_28420436 | 7.10 |
Fkbp5 |
FK506 binding protein 5 |
1819 |
0.21 |
| chr14_88428338_88428489 | 6.96 |
Gm48930 |
predicted gene, 48930 |
12388 |
0.18 |
| chr16_42998828_42999221 | 6.94 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43393 |
0.16 |
| chr17_28442321_28442494 | 6.89 |
Fkbp5 |
FK506 binding protein 5 |
685 |
0.52 |
| chr5_87499011_87499178 | 6.78 |
Ugt2a1 |
UDP glucuronosyltransferase 2 family, polypeptide A1 |
8223 |
0.12 |
| chr11_16589377_16589551 | 6.34 |
Gm12663 |
predicted gene 12663 |
46602 |
0.12 |
| chr16_93335454_93335612 | 6.29 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
17656 |
0.18 |
| chr4_141176604_141176782 | 6.21 |
Fbxo42 |
F-box protein 42 |
28771 |
0.09 |
| chr5_77102713_77103043 | 6.17 |
Hopx |
HOP homeobox |
7601 |
0.13 |
| chr6_141432376_141432556 | 6.09 |
Gm43958 |
predicted gene, 43958 |
9894 |
0.26 |
| chr4_141863008_141863177 | 5.99 |
Efhd2 |
EF hand domain containing 2 |
11828 |
0.1 |
| chr14_76149595_76149834 | 5.97 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
38823 |
0.15 |
| chr8_105122845_105122996 | 5.79 |
Ces4a |
carboxylesterase 4A |
8880 |
0.1 |
| chr4_122958854_122959026 | 5.69 |
Mfsd2a |
major facilitator superfamily domain containing 2A |
1050 |
0.41 |
| chr7_98354672_98354893 | 5.64 |
Tsku |
tsukushi, small leucine rich proteoglycan |
5297 |
0.19 |
| chr10_66930552_66930719 | 5.61 |
Gm26576 |
predicted gene, 26576 |
10333 |
0.16 |
| chr16_36933903_36934054 | 5.58 |
Hcls1 |
hematopoietic cell specific Lyn substrate 1 |
1005 |
0.38 |
| chr6_115238722_115238899 | 5.50 |
Syn2 |
synapsin II |
184 |
0.94 |
| chr12_80821404_80821588 | 5.45 |
Susd6 |
sushi domain containing 6 |
30937 |
0.11 |
| chr9_106235574_106235761 | 5.36 |
Alas1 |
aminolevulinic acid synthase 1 |
1417 |
0.25 |
| chr11_17169395_17169562 | 5.36 |
Ppp3r1 |
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
10215 |
0.17 |
| chr15_7039229_7039416 | 5.31 |
Gm38282 |
predicted gene, 38282 |
43285 |
0.17 |
| chr5_51537135_51537286 | 5.27 |
Ppargc1a |
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
16661 |
0.16 |
| chr6_108093306_108093738 | 4.94 |
Setmar |
SET domain without mariner transposase fusion |
28460 |
0.17 |
| chr1_193926856_193927030 | 4.82 |
Gm21362 |
predicted gene, 21362 |
59934 |
0.16 |
| chr17_81385422_81385814 | 4.78 |
Gm50044 |
predicted gene, 50044 |
14785 |
0.24 |
| chr4_19632141_19632303 | 4.61 |
Gm12353 |
predicted gene 12353 |
26771 |
0.17 |
| chr9_74328406_74328599 | 4.60 |
Gm24141 |
predicted gene, 24141 |
34108 |
0.17 |
| chr15_84639552_84639765 | 4.51 |
Prr5 |
proline rich 5 (renal) |
29962 |
0.16 |
| chr19_46908820_46909200 | 4.51 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
565 |
0.74 |
| chr11_38774641_38774792 | 4.51 |
Gm23520 |
predicted gene, 23520 |
23561 |
0.29 |
| chr8_105546777_105546967 | 4.47 |
Atp6v0d1 |
ATPase, H+ transporting, lysosomal V0 subunit D1 |
7396 |
0.09 |
| chr9_44078939_44079310 | 4.39 |
Usp2 |
ubiquitin specific peptidase 2 |
5815 |
0.08 |
| chr3_105758672_105758857 | 4.36 |
Rap1a |
RAS-related protein 1a |
8436 |
0.11 |
| chr17_28761138_28761314 | 4.36 |
Mapk13 |
mitogen-activated protein kinase 13 |
8071 |
0.12 |
| chr14_20148641_20148813 | 4.32 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
3433 |
0.19 |
| chr1_82188640_82188847 | 4.19 |
Gm9747 |
predicted gene 9747 |
44369 |
0.14 |
| chr1_21258115_21258471 | 4.18 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4772 |
0.12 |
| chr1_21324523_21325070 | 4.10 |
Gm21909 |
predicted gene, 21909 |
8476 |
0.1 |
| chr1_88166599_88166770 | 4.06 |
Ugt1a5 |
UDP glucuronosyltransferase 1 family, polypeptide A5 |
672 |
0.4 |
| chr2_26481863_26482061 | 3.88 |
Notch1 |
notch 1 |
11464 |
0.1 |
| chr13_101812017_101812199 | 3.80 |
Gm19108 |
predicted gene, 19108 |
30198 |
0.15 |
| chr1_74047431_74047619 | 3.78 |
Tns1 |
tensin 1 |
10368 |
0.21 |
| chr8_34975223_34975418 | 3.76 |
Gm34368 |
predicted gene, 34368 |
4554 |
0.19 |
| chr10_77440907_77441098 | 3.76 |
Gm35920 |
predicted gene, 35920 |
16915 |
0.15 |
| chr4_135337260_135337516 | 3.63 |
Srrm1 |
serine/arginine repetitive matrix 1 |
685 |
0.56 |
| chr18_12257686_12257847 | 3.62 |
Ankrd29 |
ankyrin repeat domain 29 |
13931 |
0.14 |
| chr8_22875512_22875922 | 3.52 |
Gm45555 |
predicted gene 45555 |
2128 |
0.26 |
| chr16_24782017_24782185 | 3.49 |
Mir28a |
microRNA 28a |
45754 |
0.16 |
| chr4_6239539_6239806 | 3.46 |
Gm11798 |
predicted gene 11798 |
21293 |
0.18 |
| chr16_8189232_8189397 | 3.45 |
Gm49535 |
predicted gene, 49535 |
85526 |
0.09 |
| chr3_113539787_113539957 | 3.38 |
Amy2a1 |
amylase 2a1 |
7444 |
0.22 |
| chr1_106013874_106014051 | 3.34 |
Zcchc2 |
zinc finger, CCHC domain containing 2 |
2218 |
0.27 |
| chr4_133008367_133008527 | 3.31 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
2813 |
0.24 |
| chr4_63355519_63355670 | 3.29 |
Orm3 |
orosomucoid 3 |
568 |
0.61 |
| chr10_89467947_89468098 | 3.29 |
Gas2l3 |
growth arrest-specific 2 like 3 |
24055 |
0.19 |
| chr16_46879425_46879587 | 3.26 |
Gm6912 |
predicted gene 6912 |
194692 |
0.03 |
| chr17_64760017_64760177 | 3.26 |
Gm17133 |
predicted gene 17133 |
6001 |
0.2 |
| chr17_43257833_43258015 | 3.26 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
12405 |
0.26 |
| chr13_52736343_52736501 | 3.11 |
BB123696 |
expressed sequence BB123696 |
814 |
0.75 |
| chr12_52448577_52448767 | 3.04 |
Gm47431 |
predicted gene, 47431 |
547 |
0.79 |
| chr9_24515275_24515451 | 3.03 |
Gm47772 |
predicted gene, 47772 |
11914 |
0.21 |
| chr1_75285156_75285327 | 3.01 |
Resp18 |
regulated endocrine-specific protein 18 |
6826 |
0.09 |
| chr14_17725379_17725593 | 2.96 |
Gm48320 |
predicted gene, 48320 |
45636 |
0.18 |
| chr5_147549154_147549347 | 2.91 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
9973 |
0.2 |
| chr2_181529212_181529615 | 2.89 |
Dnajc5 |
DnaJ heat shock protein family (Hsp40) member C5 |
8466 |
0.1 |
| chr7_30969422_30969656 | 2.87 |
Lsr |
lipolysis stimulated lipoprotein receptor |
3783 |
0.09 |
| chr18_12697874_12698426 | 2.80 |
Ttc39c |
tetratricopeptide repeat domain 39C |
8422 |
0.15 |
| chr15_102102548_102102719 | 2.79 |
Tns2 |
tensin 2 |
301 |
0.83 |
| chr8_122985769_122985939 | 2.79 |
Gm24445 |
predicted gene, 24445 |
2911 |
0.19 |
| chr18_74642196_74642355 | 2.79 |
Myo5b |
myosin VB |
9098 |
0.25 |
| chr12_103947018_103947616 | 2.75 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
9581 |
0.11 |
| chr4_63349571_63349895 | 2.72 |
Gm11212 |
predicted gene 11212 |
678 |
0.54 |
| chr4_63343966_63344293 | 2.68 |
Orm1 |
orosomucoid 1 |
431 |
0.73 |
| chr1_31051365_31051516 | 2.63 |
Gm28644 |
predicted gene 28644 |
9521 |
0.17 |
| chr13_31511172_31511336 | 2.60 |
Foxq1 |
forkhead box Q1 |
44880 |
0.1 |
| chr14_17699964_17700170 | 2.60 |
Thrb |
thyroid hormone receptor beta |
39171 |
0.2 |
| chr16_15889632_15889951 | 2.56 |
Cebpd |
CCAAT/enhancer binding protein (C/EBP), delta |
1375 |
0.43 |
| chr10_44700574_44700725 | 2.56 |
Gm47388 |
predicted gene, 47388 |
3469 |
0.15 |
| chr4_6937290_6937452 | 2.53 |
Tox |
thymocyte selection-associated high mobility group box |
53112 |
0.17 |
| chr2_85047151_85047302 | 2.52 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
796 |
0.47 |
| chr8_95450058_95450271 | 2.51 |
Csnk2a2 |
casein kinase 2, alpha prime polypeptide |
10550 |
0.12 |
| chr19_4493105_4493260 | 2.51 |
2010003K11Rik |
RIKEN cDNA 2010003K11 gene |
5401 |
0.13 |
| chr11_101448554_101448870 | 2.51 |
Ifi35 |
interferon-induced protein 35 |
142 |
0.87 |
| chr3_116271828_116272000 | 2.49 |
Gpr88 |
G-protein coupled receptor 88 |
18411 |
0.18 |
| chr15_100466736_100467062 | 2.47 |
5330439K02Rik |
RIKEN cDNA 5330439K02 gene |
1792 |
0.19 |
| chr3_99984293_99984453 | 2.47 |
Rpl36-ps7 |
ribosomal protein L36, pseudogene 7 |
23987 |
0.19 |
| chr7_102310929_102311109 | 2.46 |
Stim1 |
stromal interaction molecule 1 |
42692 |
0.09 |
| chr8_105104341_105104511 | 2.44 |
Gm8804 |
predicted gene 8804 |
6198 |
0.11 |
| chr1_54912808_54913219 | 2.43 |
A130048G24Rik |
RIKEN cDNA A130048G24 gene |
3581 |
0.23 |
| chr11_118249114_118249306 | 2.42 |
Cyth1 |
cytohesin 1 |
618 |
0.69 |
| chr13_30359952_30360149 | 2.38 |
Gm48161 |
predicted gene, 48161 |
10309 |
0.19 |
| chr15_81834409_81834560 | 2.36 |
Gm8444 |
predicted gene 8444 |
9229 |
0.09 |
| chr1_140407811_140407986 | 2.33 |
Kcnt2 |
potassium channel, subfamily T, member 2 |
35038 |
0.22 |
| chr2_103209491_103209661 | 2.31 |
Gm13869 |
predicted gene 13869 |
781 |
0.62 |
| chr13_103589765_103589942 | 2.26 |
Gm24870 |
predicted gene, 24870 |
52073 |
0.16 |
| chr9_48723700_48723921 | 2.25 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
112135 |
0.06 |
| chr1_164460986_164461180 | 2.24 |
Gm32391 |
predicted gene, 32391 |
1359 |
0.33 |
| chr2_30175455_30175643 | 2.23 |
Spout1 |
SPOUT domain containing methyltransferase 1 |
1956 |
0.17 |
| chr9_44086416_44086600 | 2.20 |
Usp2 |
ubiquitin specific peptidase 2 |
682 |
0.41 |
| chr15_77243081_77243232 | 2.19 |
Gm24056 |
predicted gene, 24056 |
605 |
0.67 |
| chr10_59974206_59974563 | 2.17 |
Anapc16 |
anaphase promoting complex subunit 16 |
15999 |
0.16 |
| chr2_116984707_116984867 | 2.11 |
Gm29340 |
predicted gene 29340 |
8359 |
0.2 |
| chrX_140513451_140513611 | 2.11 |
Tsc22d3 |
TSC22 domain family, member 3 |
29137 |
0.16 |
| chr7_30220091_30220286 | 2.09 |
Tbcb |
tubulin folding cofactor B |
6812 |
0.07 |
| chr2_85071072_85071223 | 2.08 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
10183 |
0.12 |
| chr1_67139611_67139949 | 2.08 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
16754 |
0.23 |
| chr8_40919479_40919678 | 2.07 |
Pdgfrl |
platelet-derived growth factor receptor-like |
6634 |
0.18 |
| chr2_165873335_165873516 | 2.04 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
2352 |
0.22 |
| chr5_135368416_135368704 | 2.03 |
Nsun5 |
NOL1/NOP2/Sun domain family, member 5 |
1393 |
0.29 |
| chr11_35693250_35693421 | 2.02 |
Pank3 |
pantothenate kinase 3 |
76149 |
0.08 |
| chr8_24429693_24429861 | 2.00 |
Gm26714 |
predicted gene, 26714 |
7373 |
0.15 |
| chr4_28646265_28646424 | 1.99 |
Gm11913 |
predicted gene 11913 |
34018 |
0.16 |
| chr10_60082039_60082224 | 1.97 |
Spock2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
24088 |
0.17 |
| chr7_121906959_121907182 | 1.92 |
Scnn1b |
sodium channel, nonvoltage-gated 1 beta |
7842 |
0.18 |
| chr11_113140051_113140245 | 1.90 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
32929 |
0.22 |
| chr17_28523067_28523337 | 1.87 |
Gm20109 |
predicted gene, 20109 |
54 |
0.53 |
| chr12_79103944_79104118 | 1.86 |
9230116L04Rik |
RIKEN cDNA 9230116L04 gene |
92 |
0.94 |
| chr10_69104650_69104828 | 1.82 |
Gm47107 |
predicted gene, 47107 |
6873 |
0.19 |
| chr10_110716862_110717029 | 1.80 |
E2f7 |
E2F transcription factor 7 |
28494 |
0.19 |
| chr19_36749089_36749269 | 1.78 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
12526 |
0.21 |
| chr17_47508103_47508259 | 1.76 |
Ccnd3 |
cyclin D3 |
2940 |
0.19 |
| chrX_97748755_97748912 | 1.71 |
Gm9062 |
predicted gene 9062 |
77546 |
0.12 |
| chr12_84316137_84316351 | 1.71 |
Zfp410 |
zinc finger protein 410 |
608 |
0.47 |
| chr15_75828201_75828376 | 1.69 |
Zc3h3 |
zinc finger CCCH type containing 3 |
79 |
0.95 |
| chr18_12734331_12734736 | 1.69 |
Ttc39c |
tetratricopeptide repeat domain 39C |
172 |
0.93 |
| chr8_123236815_123236966 | 1.69 |
Spata2l |
spermatogenesis associated 2-like |
657 |
0.4 |
| chr12_40016922_40017297 | 1.69 |
Arl4a |
ADP-ribosylation factor-like 4A |
2625 |
0.28 |
| chr11_86573938_86574127 | 1.68 |
Mir21a |
microRNA 21a |
10126 |
0.14 |
| chr4_9846537_9846689 | 1.67 |
Gdf6 |
growth differentiation factor 6 |
2241 |
0.36 |
| chr15_3359805_3359999 | 1.63 |
Ccdc152 |
coiled-coil domain containing 152 |
56376 |
0.14 |
| chr6_146501770_146501939 | 1.62 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
24 |
0.98 |
| chr6_93150084_93150947 | 1.62 |
Gm5313 |
predicted gene 5313 |
12525 |
0.22 |
| chr4_76289707_76290307 | 1.61 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
35236 |
0.22 |
| chr7_119560394_119560545 | 1.60 |
Acsm2 |
acyl-CoA synthetase medium-chain family member 2 |
1024 |
0.4 |
| chr19_3681085_3681312 | 1.59 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
5358 |
0.13 |
| chr3_81938987_81939181 | 1.58 |
Ctso |
cathepsin O |
6441 |
0.17 |
| chr13_52180080_52180254 | 1.57 |
Gm48199 |
predicted gene, 48199 |
244 |
0.95 |
| chr5_92585719_92585883 | 1.57 |
Fam47e |
family with sequence similarity 47, member E |
14245 |
0.15 |
| chr19_20650055_20650248 | 1.57 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
48190 |
0.14 |
| chr3_51373567_51373744 | 1.56 |
Gm37203 |
predicted gene, 37203 |
3433 |
0.12 |
| chr6_34863513_34863693 | 1.56 |
Tmem140 |
transmembrane protein 140 |
341 |
0.81 |
| chr3_138430446_138430657 | 1.54 |
Adh5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
12542 |
0.12 |
| chr3_97810411_97810572 | 1.54 |
Pde4dip |
phosphodiesterase 4D interacting protein (myomegalin) |
14130 |
0.2 |
| chr15_31363364_31363533 | 1.52 |
Ankrd33b |
ankyrin repeat domain 33B |
4084 |
0.18 |
| chr3_118606217_118606485 | 1.52 |
Dpyd |
dihydropyrimidine dehydrogenase |
44165 |
0.15 |
| chr5_20590310_20590514 | 1.51 |
Gm3544 |
predicted gene 3544 |
1816 |
0.41 |
| chr2_163595872_163596256 | 1.50 |
Ttpal |
tocopherol (alpha) transfer protein-like |
6250 |
0.15 |
| chr1_154117784_154118141 | 1.50 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
11 |
0.98 |
| chr1_130764110_130764467 | 1.50 |
Gm28856 |
predicted gene 28856 |
2096 |
0.18 |
| chr15_88875664_88876414 | 1.49 |
Pim3 |
proviral integration site 3 |
11119 |
0.13 |
| chr9_55289930_55290101 | 1.49 |
Nrg4 |
neuregulin 4 |
6390 |
0.18 |
| chr6_90509178_90509363 | 1.46 |
Gm44236 |
predicted gene, 44236 |
4032 |
0.14 |
| chr4_45411492_45411708 | 1.46 |
Slc25a51 |
solute carrier family 25, member 51 |
2834 |
0.21 |
| chr18_11144853_11145048 | 1.46 |
Gata6 |
GATA binding protein 6 |
85903 |
0.09 |
| chr14_32179471_32179639 | 1.44 |
Ncoa4 |
nuclear receptor coactivator 4 |
2823 |
0.15 |
| chr3_116951565_116951725 | 1.44 |
Gm42892 |
predicted gene 42892 |
7315 |
0.14 |
| chr12_12910670_12910849 | 1.42 |
Rpl36-ps3 |
ribosomal protein L36, pseudogene 3 |
1263 |
0.28 |
| chr4_125419601_125419754 | 1.42 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
71023 |
0.1 |
| chr18_56446982_56447133 | 1.40 |
Gramd3 |
GRAM domain containing 3 |
14925 |
0.19 |
| chr4_57275390_57275611 | 1.40 |
Gm12536 |
predicted gene 12536 |
24596 |
0.17 |
| chr16_91785546_91785722 | 1.40 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
1529 |
0.38 |
| chr16_4829305_4829483 | 1.40 |
4930562C15Rik |
RIKEN cDNA 4930562C15 gene |
6022 |
0.14 |
| chr19_42566701_42566858 | 1.39 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
2538 |
0.25 |
| chr16_37871477_37871675 | 1.36 |
Lrrc58 |
leucine rich repeat containing 58 |
3187 |
0.18 |
| chr4_123025711_123025964 | 1.36 |
Trit1 |
tRNA isopentenyltransferase 1 |
9240 |
0.15 |
| chr13_94441400_94441551 | 1.35 |
Ap3b1 |
adaptor-related protein complex 3, beta 1 subunit |
31388 |
0.17 |
| chr12_44270303_44270622 | 1.35 |
Pnpla8 |
patatin-like phospholipase domain containing 8 |
1295 |
0.31 |
| chr7_122074308_122074464 | 1.35 |
Ubfd1 |
ubiquitin family domain containing 1 |
5487 |
0.11 |
| chr1_67184194_67184631 | 1.34 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
61386 |
0.11 |
| chr10_43518549_43518719 | 1.34 |
Gm47889 |
predicted gene, 47889 |
63 |
0.96 |
| chr13_24144161_24144442 | 1.33 |
Carmil1 |
capping protein regulator and myosin 1 linker 1 |
10908 |
0.19 |
| chr7_119961192_119961374 | 1.33 |
Dnah3 |
dynein, axonemal, heavy chain 3 |
6559 |
0.16 |
| chr14_89799296_89799771 | 1.33 |
Gm25415 |
predicted gene, 25415 |
92728 |
0.09 |
| chr13_101719396_101719571 | 1.33 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
17379 |
0.2 |
| chr6_90464556_90464771 | 1.32 |
Klf15 |
Kruppel-like factor 15 |
641 |
0.55 |
| chr4_108348266_108348457 | 1.31 |
Coa7 |
cytochrome c oxidase assembly factor 7 |
20172 |
0.11 |
| chr11_28695120_28695290 | 1.30 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
13641 |
0.17 |
| chr16_37870060_37870244 | 1.27 |
Lrrc58 |
leucine rich repeat containing 58 |
1763 |
0.26 |
| chr8_34164335_34164526 | 1.26 |
Mir6395 |
microRNA 6395 |
2282 |
0.18 |
| chr7_81110178_81110363 | 1.25 |
Slc28a1 |
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
4529 |
0.17 |
| chr16_30964451_30964794 | 1.21 |
Gm15742 |
predicted gene 15742 |
8896 |
0.18 |
| chr7_114817644_114817801 | 1.21 |
Gm24982 |
predicted gene, 24982 |
14792 |
0.17 |
| chr11_80974682_80974867 | 1.19 |
Gm11416 |
predicted gene 11416 |
72020 |
0.1 |
| chr4_59196341_59196952 | 1.19 |
Ugcg |
UDP-glucose ceramide glucosyltransferase |
7088 |
0.18 |
| chr9_103211099_103211533 | 1.17 |
Trf |
transferrin |
690 |
0.65 |
| chr19_10167207_10167390 | 1.16 |
Gm50359 |
predicted gene, 50359 |
8863 |
0.11 |
| chr18_12667973_12668145 | 1.16 |
Gm41668 |
predicted gene, 41668 |
19640 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.6 | 3.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 3.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 0.8 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 5.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 1.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.3 | 0.5 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.3 | 1.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 0.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.2 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 2.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.7 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 1.6 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.2 | 0.9 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.4 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.2 | 0.4 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 0.6 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 0.8 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.2 | 0.9 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 0.6 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 0.4 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.2 | 2.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.2 | 0.5 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.2 | 0.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 1.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 0.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 0.7 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.5 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.2 | 0.6 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.1 | 2.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 2.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 1.3 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.4 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.7 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.1 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.1 | 0.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.4 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 | 0.5 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.7 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.3 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.1 | 0.4 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.1 | 0.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.4 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.4 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 | 0.3 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.3 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.3 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.7 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.2 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.1 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 1.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.7 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.3 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 2.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.2 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.3 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.0 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.4 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.5 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 4.9 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.2 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:1904238 | pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 1.5 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.4 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.2 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 1.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0060717 | chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.2 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.3 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:2001182 | regulation of interleukin-12 secretion(GO:2001182) |
| 0.0 | 0.1 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.2 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.4 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.0 | 0.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.2 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 1.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0001781 | neutrophil apoptotic process(GO:0001781) |
| 0.0 | 0.0 | GO:0086068 | Purkinje myocyte action potential(GO:0086017) Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:1903797 | regulation of inorganic anion transmembrane transport(GO:1903795) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.6 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.0 | GO:0060956 | endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0016045 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.9 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.0 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 1.8 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0090208 | positive regulation of triglyceride metabolic process(GO:0090208) |
| 0.0 | 0.0 | GO:0035483 | gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.0 | GO:0003139 | secondary heart field specification(GO:0003139) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.0 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0061046 | regulation of branching involved in lung morphogenesis(GO:0061046) |
| 0.0 | 0.0 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.3 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.6 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.2 | 0.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 5.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.4 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 15.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 1.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 1.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.4 | 1.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 5.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 1.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 3.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 0.2 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.1 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.1 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 2.2 | GO:0052831 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.1 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 5.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.1 | GO:0015927 | trehalase activity(GO:0015927) |
| 0.1 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 4.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 1.6 | GO:0043883 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.4 | GO:0046030 | inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0046870 | cobalt ion transmembrane transporter activity(GO:0015087) cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.1 | GO:0043908 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.2 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.8 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0044105 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0045543 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.4 | 3.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 2.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 5.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |