Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
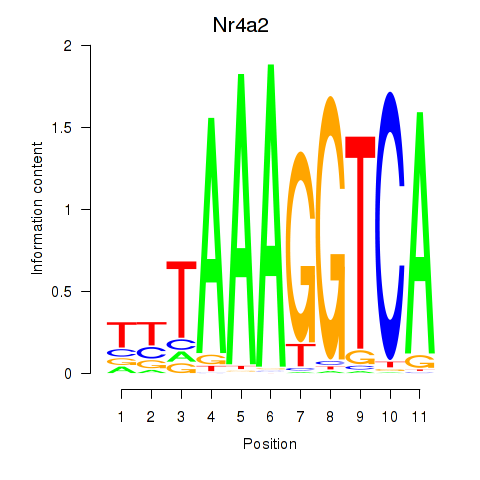
Results for Nr4a2
Z-value: 0.91
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSMUSG00000026826.7 | nuclear receptor subfamily 4, group A, member 2 |
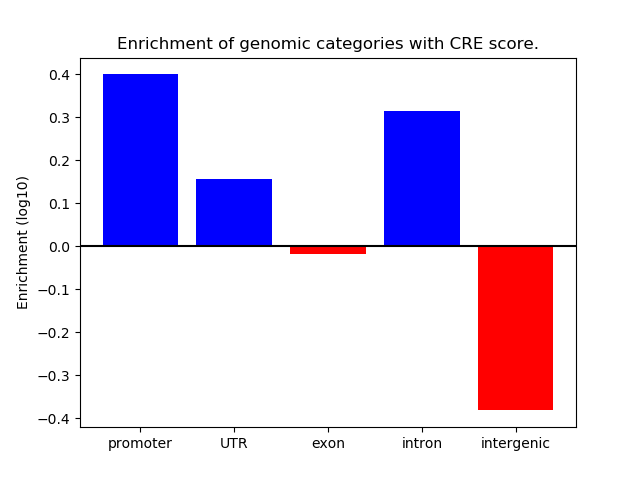
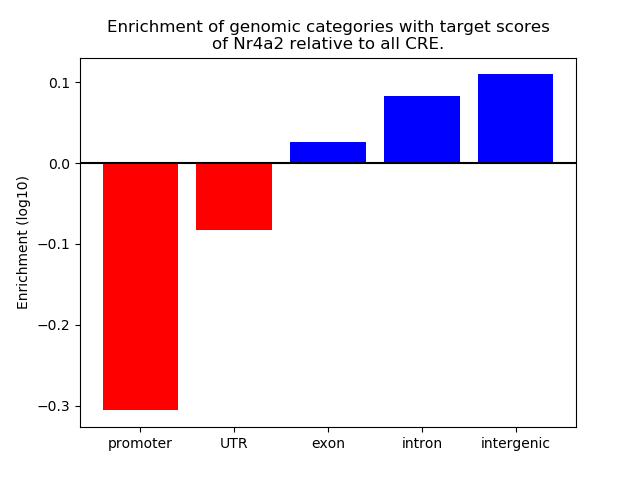
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_57117256_57117410 | Nr4a2 | 2295 | 0.311454 | -0.73 | 1.0e-01 | Click! |
| chr2_57117046_57117226 | Nr4a2 | 2098 | 0.330338 | -0.72 | 1.0e-01 | Click! |
| chr2_57127889_57128053 | Nr4a2 | 3968 | 0.231728 | -0.53 | 2.8e-01 | Click! |
| chr2_57127237_57127406 | Nr4a2 | 3318 | 0.249139 | 0.50 | 3.1e-01 | Click! |
| chr2_57117679_57117833 | Nr4a2 | 2718 | 0.281748 | 0.43 | 3.9e-01 | Click! |
Activity of the Nr4a2 motif across conditions
Conditions sorted by the z-value of the Nr4a2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
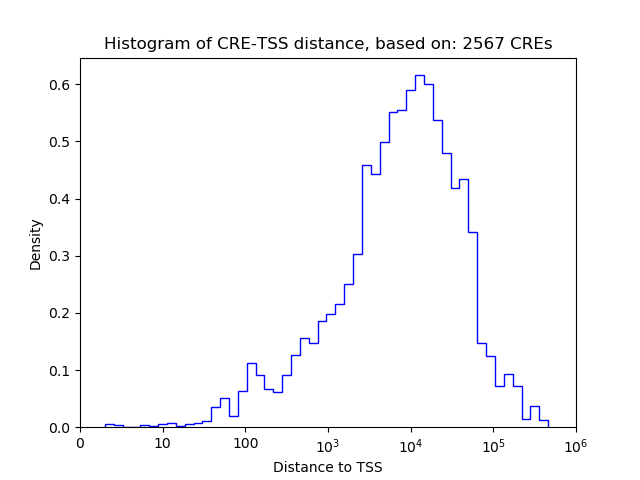
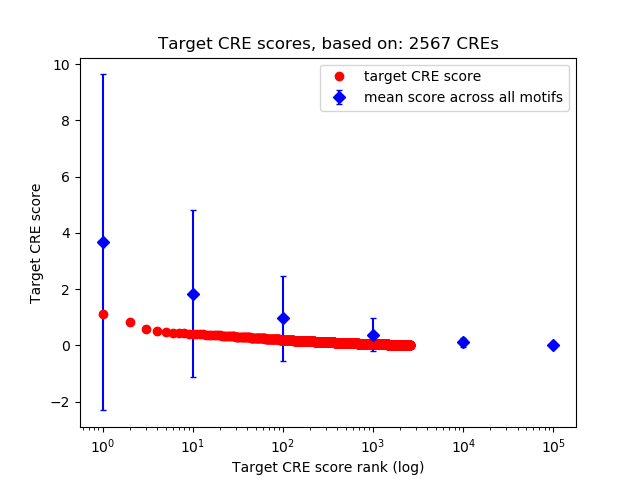
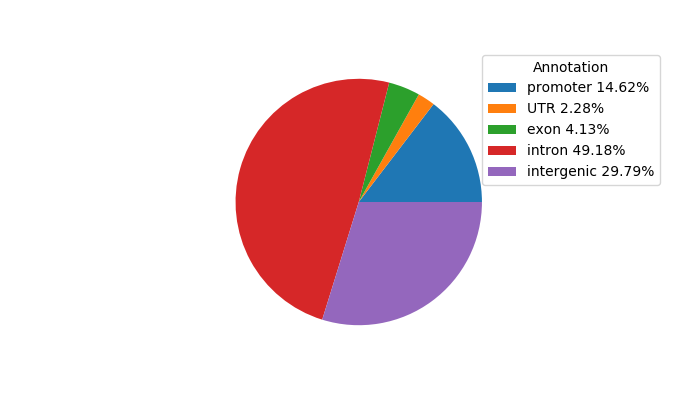
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_110381296_110381622 | 1.12 |
Gm47195 |
predicted gene, 47195 |
56074 |
0.08 |
| chr8_33950546_33950701 | 0.84 |
Rbpms |
RNA binding protein gene with multiple splicing |
20760 |
0.14 |
| chr7_143519506_143519688 | 0.58 |
Nap1l4 |
nucleosome assembly protein 1-like 4 |
5063 |
0.12 |
| chr8_112575848_112576026 | 0.50 |
Cntnap4 |
contactin associated protein-like 4 |
5882 |
0.26 |
| chr6_11309873_11310042 | 0.48 |
AA545190 |
EST AA545190 |
330319 |
0.01 |
| chr2_167351419_167351615 | 0.44 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
2334 |
0.3 |
| chr13_91771771_91771922 | 0.44 |
Acot12 |
acyl-CoA thioesterase 12 |
7482 |
0.16 |
| chr9_119405710_119405861 | 0.43 |
Gm47320 |
predicted gene, 47320 |
3231 |
0.15 |
| chr12_86817069_86817235 | 0.42 |
Gm10095 |
predicted gene 10095 |
29315 |
0.15 |
| chr9_69026149_69026327 | 0.41 |
Rora |
RAR-related orphan receptor alpha |
169303 |
0.04 |
| chr15_26779223_26779382 | 0.41 |
Gm49266 |
predicted gene, 49266 |
33165 |
0.17 |
| chr15_62137137_62137308 | 0.40 |
Pvt1 |
Pvt1 oncogene |
38193 |
0.2 |
| chr15_10330728_10330879 | 0.39 |
Gm21973 |
predicted gene 21973 |
5423 |
0.21 |
| chr7_14438643_14438794 | 0.37 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
157 |
0.95 |
| chr15_73153357_73153508 | 0.37 |
Ago2 |
argonaute RISC catalytic subunit 2 |
24415 |
0.17 |
| chr3_11357506_11357684 | 0.37 |
Gm22547 |
predicted gene, 22547 |
203258 |
0.03 |
| chr9_70138613_70138777 | 0.37 |
Fam81a |
family with sequence similarity 81, member A |
2883 |
0.24 |
| chr5_77434274_77434480 | 0.36 |
Gm34728 |
predicted gene, 34728 |
343 |
0.85 |
| chr1_140381210_140381379 | 0.35 |
Kcnt2 |
potassium channel, subfamily T, member 2 |
61642 |
0.14 |
| chr2_49720215_49720366 | 0.35 |
Kif5c |
kinesin family member 5C |
2277 |
0.35 |
| chr5_124076616_124076822 | 0.35 |
Abcb9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
2412 |
0.16 |
| chr15_84922812_84923048 | 0.34 |
Nup50 |
nucleoporin 50 |
481 |
0.69 |
| chr17_77349186_77349376 | 0.34 |
Rpsa-ps8 |
ribosomal protein SA, pseudogene 8 |
13929 |
0.31 |
| chr3_101617088_101617258 | 0.34 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
12489 |
0.17 |
| chr11_90224957_90225225 | 0.34 |
Mmd |
monocyte to macrophage differentiation-associated |
24365 |
0.15 |
| chr1_67027218_67027385 | 0.34 |
Lancl1 |
LanC (bacterial lantibiotic synthetase component C)-like 1 |
5083 |
0.22 |
| chr11_89991744_89991895 | 0.32 |
Pctp |
phosphatidylcholine transfer protein |
11075 |
0.24 |
| chrX_155248406_155248561 | 0.32 |
Gm15159 |
predicted gene 15159 |
10985 |
0.13 |
| chr1_140415820_140415994 | 0.32 |
Kcnt2 |
potassium channel, subfamily T, member 2 |
27029 |
0.25 |
| chr12_71429918_71430089 | 0.31 |
1700083H02Rik |
RIKEN cDNA 1700083H02 gene |
45795 |
0.12 |
| chr8_81634705_81634856 | 0.31 |
4930579O11Rik |
RIKEN cDNA 4930579O11 gene |
32292 |
0.22 |
| chr11_40694837_40695013 | 0.31 |
Mat2b |
methionine adenosyltransferase II, beta |
62 |
0.97 |
| chr18_52479162_52479313 | 0.31 |
Srfbp1 |
serum response factor binding protein 1 |
3858 |
0.22 |
| chr13_23592766_23592917 | 0.31 |
H4c4 |
H4 clustered histone 4 |
11243 |
0.05 |
| chrX_116629798_116629984 | 0.31 |
H2ab2 |
H2A.B variant histone 2 |
51287 |
0.17 |
| chr2_72156547_72156849 | 0.30 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
7902 |
0.2 |
| chr12_112517104_112517266 | 0.30 |
Tmem179 |
transmembrane protein 179 |
438 |
0.8 |
| chr13_69380527_69380680 | 0.29 |
Gm35514 |
predicted gene, 35514 |
14980 |
0.17 |
| chr8_56443820_56444020 | 0.29 |
Gm7419 |
predicted gene 7419 |
9595 |
0.2 |
| chr8_81366726_81366902 | 0.29 |
Inpp4b |
inositol polyphosphate-4-phosphatase, type II |
24252 |
0.21 |
| chr11_43631833_43631984 | 0.29 |
Gm12149 |
predicted gene 12149 |
15579 |
0.14 |
| chr2_122240654_122240966 | 0.28 |
Sord |
sorbitol dehydrogenase |
6061 |
0.11 |
| chr11_31778535_31778713 | 0.28 |
D630024D03Rik |
RIKEN cDNA D630024D03 gene |
45849 |
0.13 |
| chr5_72763116_72763280 | 0.27 |
Txk |
TXK tyrosine kinase |
10421 |
0.15 |
| chr13_101929962_101930142 | 0.27 |
Gm17832 |
predicted gene, 17832 |
9632 |
0.26 |
| chr2_169115164_169115359 | 0.27 |
Gm14258 |
predicted gene 14258 |
8808 |
0.23 |
| chr18_66478212_66478424 | 0.27 |
Gm50156 |
predicted gene, 50156 |
13553 |
0.11 |
| chr10_67185588_67186325 | 0.27 |
Jmjd1c |
jumonji domain containing 1C |
203 |
0.95 |
| chr16_43343299_43343463 | 0.26 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
19667 |
0.15 |
| chr14_12201682_12201851 | 0.26 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
11823 |
0.2 |
| chr3_149182211_149182384 | 0.26 |
Gm42647 |
predicted gene 42647 |
27974 |
0.19 |
| chr11_16863303_16863761 | 0.26 |
Egfr |
epidermal growth factor receptor |
14618 |
0.19 |
| chr1_136443150_136443310 | 0.26 |
Ddx59 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
10828 |
0.14 |
| chr4_150716478_150716631 | 0.26 |
Gm16079 |
predicted gene 16079 |
37762 |
0.15 |
| chr2_48442189_48442365 | 0.25 |
Gm13481 |
predicted gene 13481 |
14968 |
0.23 |
| chr13_109536856_109537007 | 0.25 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
44474 |
0.21 |
| chr10_110952952_110953127 | 0.25 |
Zdhhc17 |
zinc finger, DHHC domain containing 17 |
5342 |
0.19 |
| chr10_19491378_19491765 | 0.25 |
Gm33104 |
predicted gene, 33104 |
3072 |
0.28 |
| chr4_62723878_62724188 | 0.25 |
Gm11211 |
predicted gene 11211 |
3504 |
0.21 |
| chr12_102711766_102711920 | 0.25 |
Itpk1 |
inositol 1,3,4-triphosphate 5/6 kinase |
6913 |
0.1 |
| chr13_31180346_31180513 | 0.25 |
Gm11372 |
predicted gene 11372 |
18657 |
0.21 |
| chr4_137810112_137810287 | 0.24 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
13815 |
0.19 |
| chr11_58618424_58618625 | 0.24 |
2210407C18Rik |
RIKEN cDNA 2210407C18 gene |
2449 |
0.12 |
| chrX_60348841_60349189 | 0.24 |
Mir505 |
microRNA 505 |
45477 |
0.14 |
| chr19_29068353_29068504 | 0.24 |
Gm9895 |
predicted gene 9895 |
1081 |
0.38 |
| chr10_17299753_17299932 | 0.23 |
Gm47761 |
predicted gene, 47761 |
21769 |
0.21 |
| chr6_59072204_59072363 | 0.23 |
Gm22367 |
predicted gene, 22367 |
26970 |
0.15 |
| chr10_116753155_116753338 | 0.23 |
4930579P08Rik |
RIKEN cDNA 4930579P08 gene |
23993 |
0.15 |
| chr9_109964878_109965056 | 0.23 |
Map4 |
microtubule-associated protein 4 |
4050 |
0.14 |
| chr15_41724355_41724527 | 0.23 |
Oxr1 |
oxidation resistance 1 |
13837 |
0.26 |
| chr19_55251681_55251990 | 0.23 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
103 |
0.96 |
| chr17_14168923_14169074 | 0.23 |
Gm34510 |
predicted gene, 34510 |
34730 |
0.12 |
| chr8_18770899_18771050 | 0.23 |
Angpt2 |
angiopoietin 2 |
29412 |
0.16 |
| chr10_67097331_67097523 | 0.23 |
Reep3 |
receptor accessory protein 3 |
482 |
0.67 |
| chr1_51467935_51468124 | 0.23 |
Nabp1 |
nucleic acid binding protein 1 |
5367 |
0.2 |
| chr17_35165511_35165752 | 0.23 |
Gm17705 |
predicted gene, 17705 |
512 |
0.34 |
| chr3_12228257_12228414 | 0.23 |
Gm37996 |
predicted gene, 37996 |
156274 |
0.04 |
| chr1_105921979_105922139 | 0.23 |
Gm26214 |
predicted gene, 26214 |
23495 |
0.14 |
| chr5_111473915_111474088 | 0.22 |
Gm43119 |
predicted gene 43119 |
50412 |
0.12 |
| chr1_156838356_156838519 | 0.22 |
Angptl1 |
angiopoietin-like 1 |
125 |
0.84 |
| chr11_70845882_70846051 | 0.22 |
Rabep1 |
rabaptin, RAB GTPase binding effector protein 1 |
991 |
0.39 |
| chr12_69517687_69517891 | 0.22 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
42239 |
0.1 |
| chr9_98423674_98423967 | 0.22 |
Rbp1 |
retinol binding protein 1, cellular |
859 |
0.64 |
| chr1_23481107_23481287 | 0.22 |
Gm7784 |
predicted gene 7784 |
1658 |
0.46 |
| chr8_115983569_115983746 | 0.22 |
Gm45733 |
predicted gene 45733 |
11586 |
0.31 |
| chr17_31303465_31303616 | 0.21 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
2003 |
0.24 |
| chr17_11092974_11093138 | 0.21 |
Gm28505 |
predicted gene 28505 |
3588 |
0.3 |
| chr3_38360332_38360515 | 0.21 |
Gm42920 |
predicted gene 42920 |
8975 |
0.21 |
| chr12_70459656_70459821 | 0.21 |
Tmx1 |
thioredoxin-related transmembrane protein 1 |
103 |
0.97 |
| chr1_91104292_91104494 | 0.21 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
6821 |
0.19 |
| chr2_132638397_132638559 | 0.21 |
AU019990 |
expressed sequence AU019990 |
7403 |
0.13 |
| chr12_79762583_79762777 | 0.20 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
162053 |
0.03 |
| chr15_7160536_7160705 | 0.20 |
Lifr |
LIF receptor alpha |
6267 |
0.29 |
| chr10_41805911_41806072 | 0.20 |
Cep57l1 |
centrosomal protein 57-like 1 |
3616 |
0.19 |
| chr9_44639744_44640069 | 0.20 |
Gm47234 |
predicted gene, 47234 |
5711 |
0.08 |
| chr15_67903084_67903264 | 0.20 |
Gm49408 |
predicted gene, 49408 |
29346 |
0.21 |
| chr9_66733466_66733631 | 0.20 |
Car12 |
carbonic anhydrase 12 |
14162 |
0.17 |
| chr1_73188866_73189036 | 0.19 |
Gm38106 |
predicted gene, 38106 |
20472 |
0.23 |
| chr18_38777833_38778448 | 0.19 |
Gm8302 |
predicted gene 8302 |
5793 |
0.22 |
| chr17_26087144_26087303 | 0.19 |
Decr2 |
2-4-dienoyl-Coenzyme A reductase 2, peroxisomal |
1449 |
0.21 |
| chr13_89791634_89791792 | 0.19 |
Vcan |
versican |
49204 |
0.15 |
| chr8_91657490_91657652 | 0.19 |
Gm45295 |
predicted gene 45295 |
14326 |
0.12 |
| chr10_107964145_107964306 | 0.19 |
Gm29685 |
predicted gene, 29685 |
17580 |
0.24 |
| chr1_82168515_82168674 | 0.19 |
Mir6344 |
microRNA 6344 |
36678 |
0.16 |
| chr12_79352651_79352859 | 0.19 |
Rad51b |
RAD51 paralog B |
25402 |
0.2 |
| chr6_38737313_38737464 | 0.19 |
Hipk2 |
homeodomain interacting protein kinase 2 |
5872 |
0.25 |
| chr4_35113350_35113823 | 0.19 |
Ifnk |
interferon kappa |
38470 |
0.14 |
| chr4_152981027_152981202 | 0.18 |
Gm25779 |
predicted gene, 25779 |
32470 |
0.23 |
| chr13_93177685_93177846 | 0.18 |
Tent2 |
terminal nucleotidyltransferase 2 |
9336 |
0.2 |
| chr10_117703668_117703856 | 0.18 |
Mdm2 |
transformed mouse 3T3 cell double minute 2 |
6260 |
0.15 |
| chr1_192392344_192392537 | 0.18 |
Kcnh1 |
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
42300 |
0.19 |
| chr6_28767893_28768066 | 0.18 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
3289 |
0.27 |
| chr7_99156266_99156601 | 0.18 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
13673 |
0.12 |
| chr10_99313401_99313555 | 0.18 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
14312 |
0.14 |
| chr6_129225478_129225648 | 0.18 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
8411 |
0.12 |
| chr9_79467625_79467910 | 0.18 |
Gm10635 |
predicted gene 10635 |
51535 |
0.11 |
| chr12_111603941_111604126 | 0.18 |
Mark3 |
MAP/microtubule affinity regulating kinase 3 |
478 |
0.7 |
| chr6_93324004_93324164 | 0.18 |
Gm25094 |
predicted gene, 25094 |
11504 |
0.26 |
| chr10_99257610_99257773 | 0.18 |
Dusp6 |
dual specificity phosphatase 6 |
5540 |
0.12 |
| chr4_135859060_135859235 | 0.17 |
Srsf10 |
serine and arginine-rich splicing factor 10 |
2641 |
0.16 |
| chr3_41154447_41154774 | 0.17 |
Gm40038 |
predicted gene, 40038 |
18053 |
0.21 |
| chr5_67347155_67347319 | 0.17 |
Gm43698 |
predicted gene 43698 |
13150 |
0.16 |
| chr16_71445241_71445456 | 0.17 |
4930567J20Rik |
RIKEN cDNA 4930567J20 gene |
195464 |
0.03 |
| chr2_104813243_104813434 | 0.17 |
Qser1 |
glutamine and serine rich 1 |
3358 |
0.19 |
| chr6_88744911_88745283 | 0.17 |
Gm43999 |
predicted gene, 43999 |
2370 |
0.19 |
| chr2_119593833_119594019 | 0.17 |
Oip5os1 |
Opa interacting protein 5, opposite strand 1 |
371 |
0.78 |
| chr19_37442522_37442707 | 0.17 |
Hhex |
hematopoietically expressed homeobox |
5875 |
0.13 |
| chr3_121696912_121697098 | 0.17 |
Gm43608 |
predicted gene 43608 |
13241 |
0.11 |
| chr7_87371467_87371654 | 0.17 |
Tyr |
tyrosinase |
121832 |
0.05 |
| chr3_58659939_58660116 | 0.17 |
AU022133 |
expressed sequence AU022133 |
6539 |
0.13 |
| chr8_40573020_40573204 | 0.17 |
Mtmr7 |
myotubularin related protein 7 |
17402 |
0.16 |
| chr17_81703423_81703574 | 0.17 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
33311 |
0.22 |
| chr4_105149185_105149336 | 0.17 |
Plpp3 |
phospholipid phosphatase 3 |
8087 |
0.26 |
| chr4_118437515_118437694 | 0.17 |
Cdc20 |
cell division cycle 20 |
252 |
0.84 |
| chr17_86236900_86237066 | 0.17 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
50195 |
0.14 |
| chr3_69354794_69354993 | 0.17 |
Gm17212 |
predicted gene 17212 |
19133 |
0.17 |
| chr5_13345770_13345921 | 0.17 |
Sema3a |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
50939 |
0.16 |
| chr11_3397834_3397998 | 0.17 |
Limk2 |
LIM motif-containing protein kinase 2 |
11258 |
0.12 |
| chr13_41112373_41112548 | 0.17 |
Gcm2 |
glial cells missing homolog 2 |
1425 |
0.28 |
| chr3_157105884_157106045 | 0.17 |
Gm43526 |
predicted gene 43526 |
34220 |
0.18 |
| chr3_138256816_138256967 | 0.17 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
4100 |
0.14 |
| chr1_93439433_93439589 | 0.17 |
Hdlbp |
high density lipoprotein (HDL) binding protein |
1281 |
0.34 |
| chr10_68120874_68121044 | 0.17 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
15667 |
0.23 |
| chr3_101805513_101805698 | 0.16 |
Mab21l3 |
mab-21-like 3 |
30618 |
0.17 |
| chr4_82291564_82291726 | 0.16 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
147765 |
0.04 |
| chr8_89234098_89234358 | 0.16 |
Gm5356 |
predicted pseudogene 5356 |
46668 |
0.18 |
| chr3_81573214_81573380 | 0.16 |
Gm4857 |
predicted gene 4857 |
16472 |
0.28 |
| chr10_91261452_91261622 | 0.16 |
Gm18705 |
predicted gene, 18705 |
2193 |
0.29 |
| chr3_81289863_81290037 | 0.16 |
Gm37300 |
predicted gene, 37300 |
115622 |
0.06 |
| chr10_18067829_18067983 | 0.16 |
Reps1 |
RalBP1 associated Eps domain containing protein |
11902 |
0.2 |
| chr1_91893698_91893865 | 0.16 |
Gm37600 |
predicted gene, 37600 |
22479 |
0.17 |
| chr2_30409569_30409727 | 0.16 |
Crat |
carnitine acetyltransferase |
2969 |
0.13 |
| chr15_27913676_27913865 | 0.16 |
Trio |
triple functional domain (PTPRF interacting) |
5501 |
0.24 |
| chr15_81915764_81916002 | 0.16 |
Polr3h |
polymerase (RNA) III (DNA directed) polypeptide H |
754 |
0.45 |
| chr8_45432180_45432331 | 0.16 |
Tlr3 |
toll-like receptor 3 |
21175 |
0.15 |
| chr7_25450039_25450206 | 0.16 |
Gm15495 |
predicted gene 15495 |
6659 |
0.11 |
| chr1_171212888_171213039 | 0.16 |
Nr1i3 |
nuclear receptor subfamily 1, group I, member 3 |
1007 |
0.29 |
| chr10_17667448_17667608 | 0.16 |
Gm47771 |
predicted gene, 47771 |
9015 |
0.23 |
| chr11_68540853_68541004 | 0.16 |
Pik3r6 |
phosphoinositide-3-kinase regulatory subunit 5 |
4728 |
0.19 |
| chr15_6882256_6882435 | 0.16 |
Osmr |
oncostatin M receptor |
7376 |
0.29 |
| chr3_89436835_89437052 | 0.16 |
Pbxip1 |
pre B cell leukemia transcription factor interacting protein 1 |
187 |
0.87 |
| chrX_129167854_129168016 | 0.16 |
Gm26029 |
predicted gene, 26029 |
11050 |
0.3 |
| chr19_53889732_53889907 | 0.15 |
Pdcd4 |
programmed cell death 4 |
2412 |
0.23 |
| chr2_4941693_4941844 | 0.15 |
Gm13194 |
predicted gene 13194 |
406 |
0.76 |
| chr15_31251286_31251565 | 0.15 |
Dap |
death-associated protein |
16911 |
0.15 |
| chr6_87805806_87806031 | 0.15 |
9930120I10Rik |
RIKEN cDNA 9930120I10 gene |
418 |
0.64 |
| chr6_49210657_49210827 | 0.15 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
3419 |
0.2 |
| chr7_16046893_16047053 | 0.15 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
948 |
0.46 |
| chr2_24183784_24184156 | 0.15 |
Il1f9 |
interleukin 1 family, member 9 |
2506 |
0.2 |
| chr2_92943609_92943778 | 0.15 |
Syt13 |
synaptotagmin XIII |
28595 |
0.15 |
| chr4_132655416_132655575 | 0.15 |
Eya3 |
EYA transcriptional coactivator and phosphatase 3 |
1205 |
0.44 |
| chr4_107388361_107388798 | 0.15 |
Ndc1 |
NDC1 transmembrane nucleoporin |
7117 |
0.15 |
| chr14_27679797_27679980 | 0.15 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
27061 |
0.19 |
| chr4_108066732_108066892 | 0.15 |
Scp2 |
sterol carrier protein 2, liver |
4551 |
0.16 |
| chr2_5219339_5219504 | 0.15 |
Gm23118 |
predicted gene, 23118 |
40694 |
0.14 |
| chr10_128921362_128921572 | 0.15 |
Rdh5 |
retinol dehydrogenase 5 |
1421 |
0.19 |
| chr4_124670340_124670505 | 0.15 |
Gm2164 |
predicted gene 2164 |
13253 |
0.09 |
| chr16_97497728_97497915 | 0.15 |
Gm24777 |
predicted gene, 24777 |
1201 |
0.36 |
| chr5_114122907_114123086 | 0.15 |
Alkbh2 |
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
5180 |
0.11 |
| chr6_120589309_120589479 | 0.15 |
Gm44124 |
predicted gene, 44124 |
9218 |
0.14 |
| chr8_46404315_46404484 | 0.15 |
Gm45253 |
predicted gene 45253 |
18511 |
0.13 |
| chr6_87427046_87427210 | 0.15 |
Bmp10 |
bone morphogenetic protein 10 |
1866 |
0.27 |
| chr16_10347675_10347844 | 0.15 |
Gm1600 |
predicted gene 1600 |
168 |
0.94 |
| chr6_94550970_94551136 | 0.15 |
Slc25a26 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
16913 |
0.19 |
| chr14_48072185_48072471 | 0.15 |
Gm6055 |
predicted gene 6055 |
7424 |
0.16 |
| chr18_56974912_56975237 | 0.15 |
C330018D20Rik |
RIKEN cDNA C330018D20 gene |
172 |
0.96 |
| chr15_34477411_34477563 | 0.15 |
Gm49188 |
predicted gene, 49188 |
4220 |
0.13 |
| chr8_89097725_89097891 | 0.15 |
Gm6625 |
predicted gene 6625 |
49250 |
0.14 |
| chr6_5223673_5223834 | 0.15 |
Gm44250 |
predicted gene, 44250 |
3267 |
0.22 |
| chr5_25054101_25054285 | 0.15 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
46449 |
0.11 |
| chr6_108435674_108435848 | 0.15 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
17884 |
0.19 |
| chr4_44776736_44776900 | 0.15 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
16374 |
0.16 |
| chr10_18370393_18370778 | 0.15 |
Nhsl1 |
NHS-like 1 |
19737 |
0.19 |
| chr4_148602965_148603132 | 0.14 |
Masp2 |
mannan-binding lectin serine peptidase 2 |
473 |
0.67 |
| chr13_108719037_108719229 | 0.14 |
Rps3a3 |
ribosomal protein S3A3 |
48530 |
0.17 |
| chr6_142894834_142895115 | 0.14 |
St8sia1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
11934 |
0.19 |
| chr10_87704249_87704407 | 0.14 |
Gm48195 |
predicted gene, 48195 |
15750 |
0.25 |
| chr6_120190995_120191345 | 0.14 |
Ninj2 |
ninjurin 2 |
2653 |
0.29 |
| chr10_61303355_61303525 | 0.14 |
Prf1 |
perforin 1 (pore forming protein) |
3639 |
0.17 |
| chr1_128158652_128158822 | 0.14 |
Gm37407 |
predicted gene, 37407 |
4477 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.0 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |