Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
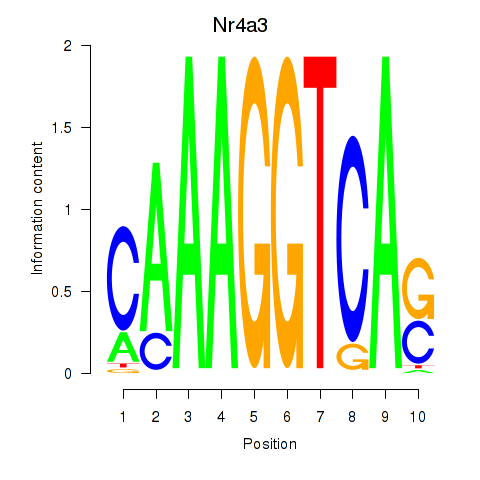
Results for Nr4a3
Z-value: 2.30
Transcription factors associated with Nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a3
|
ENSMUSG00000028341.3 | nuclear receptor subfamily 4, group A, member 3 |
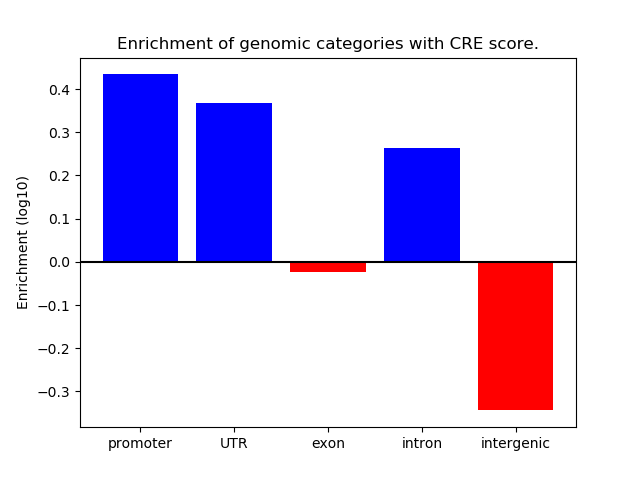
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_48042684_48042844 | Nr4a3 | 2389 | 0.338726 | 0.94 | 6.0e-03 | Click! |
| chr4_48046634_48046887 | Nr4a3 | 1529 | 0.456651 | 0.88 | 2.1e-02 | Click! |
| chr4_48044008_48044162 | Nr4a3 | 1068 | 0.583345 | 0.85 | 3.2e-02 | Click! |
| chr4_48045019_48045175 | Nr4a3 | 56 | 0.980703 | 0.80 | 5.6e-02 | Click! |
| chr4_48045892_48046043 | Nr4a3 | 736 | 0.721413 | 0.73 | 1.0e-01 | Click! |
Activity of the Nr4a3 motif across conditions
Conditions sorted by the z-value of the Nr4a3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
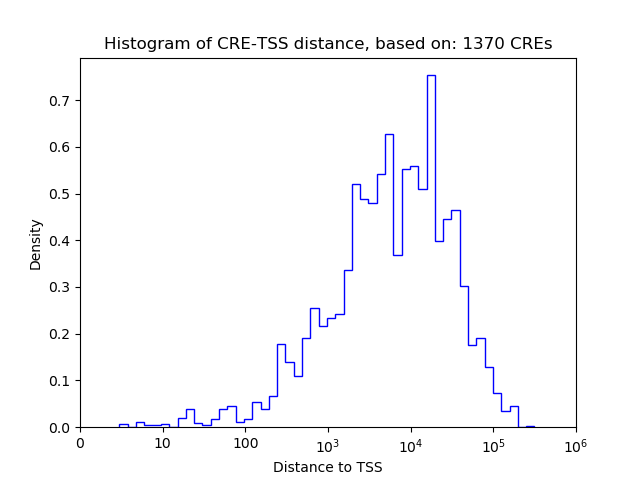
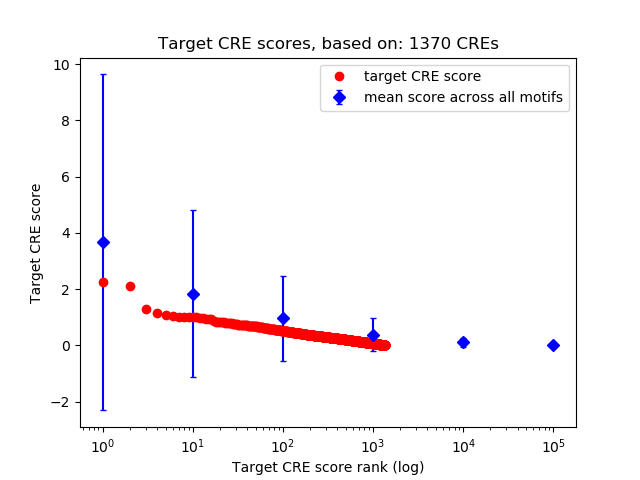
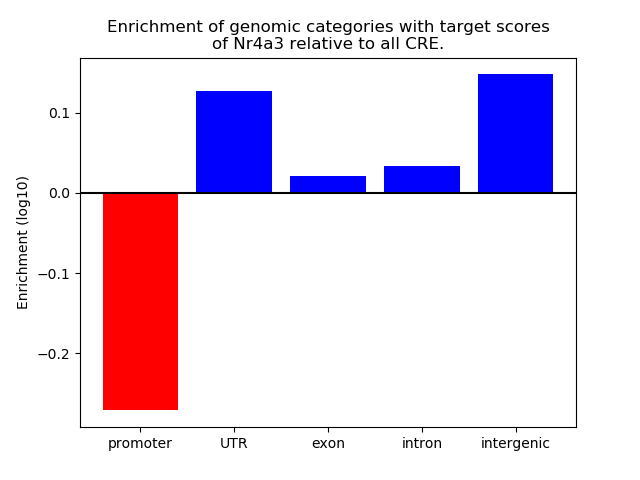
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_125976536_125976742 | 2.25 |
Kcnk1 |
potassium channel, subfamily K, member 1 |
18531 |
0.19 |
| chr15_25435984_25436493 | 2.10 |
Gm48956 |
predicted gene, 48956 |
9402 |
0.16 |
| chr10_99600138_99600289 | 1.28 |
Gm20110 |
predicted gene, 20110 |
8958 |
0.19 |
| chr8_45691473_45691656 | 1.14 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
3227 |
0.28 |
| chr8_123978504_123978907 | 1.09 |
Abcb10 |
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
4417 |
0.12 |
| chr6_119487729_119487894 | 1.05 |
Fbxl14 |
F-box and leucine-rich repeat protein 14 |
8143 |
0.21 |
| chr8_24506282_24506451 | 1.02 |
9130214F15Rik |
RIKEN cDNA 9130214F15 gene |
64 |
0.97 |
| chr5_93296317_93296483 | 1.02 |
Ccng2 |
cyclin G2 |
27692 |
0.16 |
| chr17_81389463_81389664 | 1.01 |
Gm50044 |
predicted gene, 50044 |
18730 |
0.23 |
| chr4_129191808_129191992 | 1.00 |
Yars |
tyrosyl-tRNA synthetase |
1998 |
0.2 |
| chr1_187313973_187314152 | 0.99 |
Gm38155 |
predicted gene, 38155 |
63407 |
0.1 |
| chr19_3650560_3650722 | 0.98 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
34849 |
0.1 |
| chr1_79466047_79466371 | 0.96 |
Scg2 |
secretogranin II |
26089 |
0.18 |
| chr13_54583316_54583476 | 0.95 |
Arl10 |
ADP-ribosylation factor-like 10 |
4584 |
0.11 |
| chr9_122018172_122018323 | 0.94 |
Gm47117 |
predicted gene, 47117 |
6107 |
0.11 |
| chr3_100536474_100536643 | 0.94 |
Gm42868 |
predicted gene 42868 |
39129 |
0.11 |
| chr3_52646413_52646568 | 0.88 |
Gm10293 |
predicted pseudogene 10293 |
33655 |
0.18 |
| chr16_34746212_34746363 | 0.82 |
Mylk |
myosin, light polypeptide kinase |
1077 |
0.6 |
| chr12_26387378_26387859 | 0.82 |
Rnf144a |
ring finger protein 144A |
18829 |
0.15 |
| chr5_145864595_145865130 | 0.81 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
11829 |
0.16 |
| chr4_134722177_134722332 | 0.81 |
Man1c1 |
mannosidase, alpha, class 1C, member 1 |
17964 |
0.18 |
| chr14_51289185_51289366 | 0.81 |
Gm49245 |
predicted gene, 49245 |
11673 |
0.1 |
| chr8_117637336_117637505 | 0.81 |
Sdr42e1 |
short chain dehydrogenase/reductase family 42E, member 1 |
34127 |
0.12 |
| chr6_6183421_6183599 | 0.80 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
7809 |
0.26 |
| chr9_65191450_65191617 | 0.80 |
Parp16 |
poly (ADP-ribose) polymerase family, member 16 |
3820 |
0.14 |
| chr2_167877367_167877523 | 0.80 |
Gm14319 |
predicted gene 14319 |
18860 |
0.16 |
| chr1_179632334_179632515 | 0.78 |
Sccpdh |
saccharopine dehydrogenase (putative) |
35786 |
0.14 |
| chr8_127515714_127515885 | 0.76 |
Gm6921 |
predicted pseudogene 6921 |
45636 |
0.17 |
| chr2_163598624_163598794 | 0.76 |
Ttpal |
tocopherol (alpha) transfer protein-like |
3605 |
0.18 |
| chrX_85053601_85053753 | 0.75 |
Dmd |
dystrophin, muscular dystrophy |
5264 |
0.33 |
| chr10_75033725_75033908 | 0.74 |
Gm40692 |
predicted gene, 40692 |
529 |
0.63 |
| chr3_85273227_85273668 | 0.73 |
1700036G14Rik |
RIKEN cDNA 1700036G14 gene |
44072 |
0.16 |
| chr5_56910070_56910250 | 0.73 |
Gm23896 |
predicted gene, 23896 |
174070 |
0.03 |
| chr18_20936854_20937178 | 0.73 |
Rnf125 |
ring finger protein 125 |
7609 |
0.22 |
| chr10_42194110_42194261 | 0.72 |
Foxo3 |
forkhead box O3 |
64181 |
0.12 |
| chr16_79059250_79059443 | 0.72 |
Tmprss15 |
transmembrane protease, serine 15 |
31746 |
0.22 |
| chr12_29257874_29258025 | 0.72 |
Gm6989 |
predicted gene 6989 |
59619 |
0.14 |
| chr6_62934342_62934493 | 0.72 |
Gm5001 |
predicted gene 5001 |
5271 |
0.31 |
| chr7_24504957_24505109 | 0.71 |
Zfp428 |
zinc finger protein 428 |
1973 |
0.15 |
| chr4_145279031_145279214 | 0.71 |
Tnfrsf1b |
tumor necrosis factor receptor superfamily, member 1b |
32252 |
0.14 |
| chr2_163616887_163617081 | 0.70 |
Ttpal |
tocopherol (alpha) transfer protein-like |
5464 |
0.15 |
| chr10_60733446_60733765 | 0.69 |
Slc29a3 |
solute carrier family 29 (nucleoside transporters), member 3 |
9289 |
0.21 |
| chr14_19710131_19710282 | 0.69 |
Gm49341 |
predicted gene, 49341 |
12997 |
0.13 |
| chr4_135746130_135746372 | 0.69 |
Gm12988 |
predicted gene 12988 |
10444 |
0.12 |
| chr1_36525855_36526032 | 0.69 |
Gm38033 |
predicted gene, 38033 |
2294 |
0.14 |
| chr10_84907603_84907783 | 0.69 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
9923 |
0.24 |
| chr14_66521966_66522122 | 0.68 |
Gm23899 |
predicted gene, 23899 |
69651 |
0.11 |
| chr10_79616142_79616293 | 0.68 |
C2cd4c |
C2 calcium-dependent domain containing 4C |
2192 |
0.15 |
| chr1_51996762_51997232 | 0.68 |
Stat4 |
signal transducer and activator of transcription 4 |
9849 |
0.17 |
| chr5_90894238_90894431 | 0.67 |
Gm22816 |
predicted gene, 22816 |
954 |
0.41 |
| chr14_25295396_25295547 | 0.67 |
Gm26660 |
predicted gene, 26660 |
83164 |
0.08 |
| chr19_30171407_30171581 | 0.67 |
Gldc |
glycine decarboxylase |
3935 |
0.23 |
| chr18_15330500_15330684 | 0.65 |
E430002N23Rik |
RIKEN cDNA E430002N23 gene |
17893 |
0.18 |
| chr2_166637694_166637887 | 0.65 |
Prex1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
1178 |
0.52 |
| chr17_45322333_45322491 | 0.64 |
Cdc5l |
cell division cycle 5-like (S. pombe) |
93272 |
0.06 |
| chr8_25819139_25819316 | 0.64 |
Ash2l |
ASH2 like histone lysine methyltransferase complex subunit |
4348 |
0.13 |
| chr5_76297411_76297591 | 0.64 |
Gm43658 |
predicted gene 43658 |
4274 |
0.14 |
| chr2_33369566_33369732 | 0.64 |
Ralgps1 |
Ral GEF with PH domain and SH3 binding motif 1 |
1780 |
0.3 |
| chr19_5880801_5880961 | 0.64 |
Slc25a45 |
solute carrier family 25, member 45 |
2225 |
0.13 |
| chr3_18463182_18463725 | 0.63 |
Gm30667 |
predicted gene, 30667 |
2199 |
0.33 |
| chr1_133933859_133934320 | 0.62 |
Prelp |
proline arginine-rich end leucine-rich repeat |
12675 |
0.13 |
| chr15_102177030_102177199 | 0.62 |
Csad |
cysteine sulfinic acid decarboxylase |
1905 |
0.2 |
| chr10_28166717_28166892 | 0.61 |
Gm22370 |
predicted gene, 22370 |
47317 |
0.17 |
| chr8_103417794_103417953 | 0.61 |
1600027J07Rik |
RIKEN cDNA 1600027J07 gene |
70339 |
0.12 |
| chr13_114974629_114975014 | 0.61 |
Gm47776 |
predicted gene, 47776 |
39753 |
0.15 |
| chr13_56477354_56477672 | 0.60 |
Il9 |
interleukin 9 |
4733 |
0.19 |
| chr6_136469608_136469798 | 0.60 |
Gm6728 |
predicted gene 6728 |
17483 |
0.12 |
| chr11_5917118_5917412 | 0.60 |
Gm11967 |
predicted gene 11967 |
1950 |
0.2 |
| chr1_134293960_134294142 | 0.60 |
Myog |
myogenin |
4062 |
0.15 |
| chr1_155158041_155158192 | 0.60 |
Stx6 |
syntaxin 6 |
599 |
0.68 |
| chr15_96031527_96031698 | 0.59 |
D030018L15Rik |
RIKEN cDNA D030018L15 gene |
23213 |
0.19 |
| chr16_91833643_91834213 | 0.59 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
5771 |
0.19 |
| chr12_41208691_41208865 | 0.59 |
Gm47376 |
predicted gene, 47376 |
139312 |
0.04 |
| chr19_4700355_4700506 | 0.59 |
Gm960 |
predicted gene 960 |
1762 |
0.22 |
| chr16_95673938_95674224 | 0.58 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
27994 |
0.19 |
| chr7_109599227_109599378 | 0.58 |
Denn2b |
DENN domain containing 2B |
3363 |
0.22 |
| chr10_81417531_81417694 | 0.57 |
Mir1191b |
microRNA 1191b |
1315 |
0.18 |
| chr1_130763367_130763536 | 0.57 |
Gm28856 |
predicted gene 28856 |
2933 |
0.14 |
| chr8_84069298_84069478 | 0.57 |
C330011M18Rik |
RIKEN cDNA C330011M18 gene |
2101 |
0.12 |
| chr5_92134368_92134534 | 0.56 |
Uso1 |
USO1 vesicle docking factor |
3487 |
0.15 |
| chr19_5567308_5567477 | 0.56 |
Ap5b1 |
adaptor-related protein complex 5, beta 1 subunit |
633 |
0.46 |
| chr18_31966444_31966632 | 0.56 |
Lims2 |
LIM and senescent cell antigen like domains 2 |
10205 |
0.13 |
| chr2_118883051_118883401 | 0.55 |
Ivd |
isovaleryl coenzyme A dehydrogenase |
6793 |
0.14 |
| chr9_77847197_77847348 | 0.55 |
Gm19572 |
predicted gene, 19572 |
4436 |
0.16 |
| chr11_26592820_26593046 | 0.54 |
Gm6899 |
predicted gene 6899 |
293 |
0.76 |
| chr2_167799424_167799897 | 0.54 |
9230111E07Rik |
RIKEN cDNA 9230111E07 gene |
17607 |
0.14 |
| chr1_73901414_73901565 | 0.54 |
Tns1 |
tensin 1 |
26728 |
0.15 |
| chr1_156074597_156074881 | 0.54 |
Tor1aip2 |
torsin A interacting protein 2 |
11526 |
0.17 |
| chr13_14191910_14192117 | 0.54 |
Arid4b |
AT rich interactive domain 4B (RBP1-like) |
836 |
0.59 |
| chr5_115475861_115476388 | 0.54 |
Sirt4 |
sirtuin 4 |
3814 |
0.1 |
| chr8_25750561_25750717 | 0.54 |
Ddhd2 |
DDHD domain containing 2 |
789 |
0.44 |
| chr12_59191626_59191786 | 0.53 |
Mia2 |
MIA SH3 domain ER export factor 2 |
12193 |
0.14 |
| chr5_140171802_140172150 | 0.53 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
2683 |
0.24 |
| chr11_97836691_97836945 | 0.53 |
Lasp1 |
LIM and SH3 protein 1 |
3394 |
0.12 |
| chr13_43252482_43252652 | 0.53 |
Gfod1 |
glucose-fructose oxidoreductase domain containing 1 |
50838 |
0.13 |
| chr4_124737570_124737747 | 0.52 |
Gm24721 |
predicted gene, 24721 |
1819 |
0.16 |
| chr10_43487752_43488061 | 0.52 |
Bend3 |
BEN domain containing 3 |
2672 |
0.19 |
| chr11_117780625_117780787 | 0.52 |
Tmc6 |
transmembrane channel-like gene family 6 |
48 |
0.93 |
| chr4_150732960_150733140 | 0.52 |
Gm16079 |
predicted gene 16079 |
54258 |
0.11 |
| chr19_6251158_6251334 | 0.52 |
Atg2a |
autophagy related 2A |
242 |
0.81 |
| chr1_62709908_62710059 | 0.52 |
Nrp2 |
neuropilin 2 |
6281 |
0.18 |
| chr13_54439539_54439690 | 0.52 |
Thoc3 |
THO complex 3 |
29174 |
0.12 |
| chr18_35256811_35256978 | 0.51 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
4573 |
0.19 |
| chr9_111440393_111440743 | 0.51 |
Dclk3 |
doublecortin-like kinase 3 |
1487 |
0.42 |
| chr16_3843876_3844135 | 0.51 |
Zfp174 |
zinc finger protein 174 |
3263 |
0.13 |
| chr11_83850528_83850683 | 0.51 |
Hnf1b |
HNF1 homeobox B |
250 |
0.85 |
| chr2_160805166_160805505 | 0.51 |
Gm11447 |
predicted gene 11447 |
40278 |
0.11 |
| chr11_78972853_78973033 | 0.51 |
Lgals9 |
lectin, galactose binding, soluble 9 |
2613 |
0.27 |
| chr1_153307781_153308161 | 0.50 |
Gm8818 |
predicted pseudogene 8818 |
2539 |
0.25 |
| chr2_25708935_25709097 | 0.49 |
A230005M16Rik |
RIKEN cDNA A230005M16 gene |
700 |
0.42 |
| chr4_144957863_144958036 | 0.49 |
Gm38074 |
predicted gene, 38074 |
899 |
0.6 |
| chr19_33098081_33098289 | 0.49 |
Gm29946 |
predicted gene, 29946 |
24462 |
0.18 |
| chr19_40218990_40219184 | 0.49 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
4269 |
0.17 |
| chr2_43513507_43513674 | 0.48 |
Gm13464 |
predicted gene 13464 |
5808 |
0.3 |
| chr6_39796421_39796591 | 0.48 |
Mrps33 |
mitochondrial ribosomal protein S33 |
9536 |
0.17 |
| chr2_166678201_166678352 | 0.48 |
Gm23152 |
predicted gene, 23152 |
23134 |
0.17 |
| chr10_84477716_84477877 | 0.48 |
4930463O16Rik |
RIKEN cDNA 4930463O16 gene |
10497 |
0.12 |
| chr19_38709785_38709951 | 0.47 |
Gm8717 |
predicted gene 8717 |
5544 |
0.21 |
| chr11_88574733_88574900 | 0.47 |
Msi2 |
musashi RNA-binding protein 2 |
15331 |
0.24 |
| chrX_160424988_160425139 | 0.47 |
Adgrg2 |
adhesion G protein-coupled receptor G2 |
2229 |
0.34 |
| chr17_26702508_26702992 | 0.47 |
Crebrf |
CREB3 regulatory factor |
12900 |
0.14 |
| chr3_52469938_52470147 | 0.47 |
Gm38098 |
predicted gene, 38098 |
44634 |
0.14 |
| chr3_69543290_69543467 | 0.47 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
946 |
0.58 |
| chr18_4956609_4956805 | 0.47 |
Svil |
supervillin |
34981 |
0.22 |
| chr14_100224229_100224566 | 0.46 |
Gm16260 |
predicted gene 16260 |
4911 |
0.22 |
| chr4_154247269_154247441 | 0.46 |
Megf6 |
multiple EGF-like-domains 6 |
1941 |
0.28 |
| chr1_121321522_121321673 | 0.46 |
Insig2 |
insulin induced gene 2 |
1594 |
0.33 |
| chr1_157084475_157084641 | 0.46 |
Tex35 |
testis expressed 35 |
23830 |
0.14 |
| chr6_71989276_71989427 | 0.46 |
Gm26628 |
predicted gene, 26628 |
25576 |
0.11 |
| chr13_46161900_46162051 | 0.46 |
Gm10113 |
predicted gene 10113 |
29071 |
0.21 |
| chr3_10282975_10283159 | 0.45 |
Fabp12 |
fatty acid binding protein 12 |
18107 |
0.09 |
| chr3_104568490_104568648 | 0.45 |
Gm26091 |
predicted gene, 26091 |
32670 |
0.1 |
| chr1_118448989_118449484 | 0.45 |
Gm26080 |
predicted gene, 26080 |
4249 |
0.15 |
| chr2_34486208_34486368 | 0.45 |
Mapkap1 |
mitogen-activated protein kinase associated protein 1 |
41941 |
0.13 |
| chr1_91899152_91899309 | 0.45 |
Gm37600 |
predicted gene, 37600 |
17030 |
0.18 |
| chr1_91095685_91095922 | 0.45 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
15411 |
0.18 |
| chr1_20618287_20618471 | 0.45 |
Pkhd1 |
polycystic kidney and hepatic disease 1 |
315 |
0.9 |
| chr2_173162438_173162602 | 0.44 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
9438 |
0.17 |
| chr7_102132512_102132682 | 0.44 |
Chrna10 |
cholinergic receptor, nicotinic, alpha polypeptide 10 |
15769 |
0.1 |
| chr12_80126455_80126626 | 0.44 |
2310015A10Rik |
RIKEN cDNA 2310015A10 gene |
5401 |
0.14 |
| chr4_15327477_15327645 | 0.44 |
Gm24317 |
predicted gene, 24317 |
55587 |
0.13 |
| chr19_10041963_10042141 | 0.44 |
Fads3 |
fatty acid desaturase 3 |
320 |
0.83 |
| chr6_119681588_119681760 | 0.43 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
59569 |
0.13 |
| chr6_72604103_72604254 | 0.43 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
195 |
0.84 |
| chr11_46084943_46085145 | 0.43 |
Mir8100 |
microRNA 8100 |
17226 |
0.12 |
| chr16_10981270_10981624 | 0.43 |
Litaf |
LPS-induced TN factor |
5868 |
0.12 |
| chr7_134192391_134192592 | 0.43 |
Adam12 |
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
19547 |
0.21 |
| chr17_68285429_68285617 | 0.43 |
L3mbtl4 |
L3MBTL4 histone methyl-lysine binding protein |
11726 |
0.26 |
| chr17_65931229_65931442 | 0.43 |
Twsg1 |
twisted gastrulation BMP signaling modulator 1 |
4926 |
0.15 |
| chr3_100189024_100189191 | 0.43 |
Gdap2 |
ganglioside-induced differentiation-associated-protein 2 |
5473 |
0.23 |
| chr9_108858778_108859092 | 0.43 |
Slc26a6 |
solute carrier family 26, member 6 |
3273 |
0.12 |
| chr1_59157472_59157626 | 0.43 |
Mpp4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
1112 |
0.34 |
| chr1_182125404_182125574 | 0.43 |
Srp9 |
signal recognition particle 9 |
708 |
0.61 |
| chr6_99666342_99666493 | 0.43 |
Eif4e3 |
eukaryotic translation initiation factor 4E member 3 |
354 |
0.48 |
| chr3_145561026_145561180 | 0.42 |
Znhit6 |
zinc finger, HIT type 6 |
15102 |
0.19 |
| chr4_136018964_136019115 | 0.42 |
Gm24362 |
predicted gene, 24362 |
553 |
0.57 |
| chr7_100077379_100077530 | 0.42 |
Pold3 |
polymerase (DNA-directed), delta 3, accessory subunit |
12583 |
0.16 |
| chr18_57029725_57029909 | 0.42 |
C330018D20Rik |
RIKEN cDNA C330018D20 gene |
54449 |
0.12 |
| chr5_140107803_140107986 | 0.41 |
Gm16121 |
predicted gene 16121 |
6695 |
0.16 |
| chr4_89526966_89527117 | 0.41 |
Gm12608 |
predicted gene 12608 |
82397 |
0.09 |
| chr11_94158519_94158921 | 0.41 |
B230206L02Rik |
RIKEN cDNA B230206L02 gene |
23060 |
0.17 |
| chr10_59784742_59784904 | 0.41 |
Gm17059 |
predicted gene 17059 |
15431 |
0.14 |
| chr3_130692655_130693111 | 0.41 |
Ostc |
oligosaccharyltransferase complex subunit (non-catalytic) |
3739 |
0.17 |
| chr7_45973330_45973912 | 0.41 |
Abcc6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
302 |
0.76 |
| chr2_169075125_169075304 | 0.40 |
Gm26469 |
predicted gene, 26469 |
14094 |
0.19 |
| chr15_88830094_88830418 | 0.40 |
Gm23144 |
predicted gene, 23144 |
5044 |
0.15 |
| chr12_29136516_29136667 | 0.40 |
Gm31333 |
predicted gene, 31333 |
2576 |
0.33 |
| chr4_145054462_145054637 | 0.40 |
Vps13d |
vacuolar protein sorting 13D |
406 |
0.89 |
| chr6_17469322_17469539 | 0.40 |
Met |
met proto-oncogene |
5323 |
0.23 |
| chr9_46238922_46239117 | 0.40 |
Apoa4 |
apolipoprotein A-IV |
1677 |
0.17 |
| chr10_99207689_99207863 | 0.40 |
Poc1b |
POC1 centriolar protein B |
14884 |
0.11 |
| chr17_11752805_11752972 | 0.40 |
Gm10513 |
predicted gene 10513 |
20543 |
0.26 |
| chr15_81337946_81338137 | 0.40 |
Slc25a17 |
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
6116 |
0.16 |
| chr7_136883017_136883173 | 0.40 |
Mgmt |
O-6-methylguanine-DNA methyltransferase |
11519 |
0.23 |
| chr9_88355859_88356010 | 0.40 |
Nt5e |
5' nucleotidase, ecto |
247 |
0.9 |
| chr6_37592493_37592657 | 0.40 |
Gm7463 |
predicted gene 7463 |
51161 |
0.14 |
| chrX_7766214_7766424 | 0.39 |
Tfe3 |
transcription factor E3 |
1667 |
0.18 |
| chr8_10914649_10914809 | 0.39 |
3930402G23Rik |
RIKEN cDNA 3930402G23 gene |
13715 |
0.09 |
| chr15_82165521_82165825 | 0.39 |
Srebf2 |
sterol regulatory element binding factor 2 |
17648 |
0.09 |
| chr16_23108839_23109021 | 0.39 |
Snord2 |
small nucleolar RNA, C/D box 2 |
23 |
0.76 |
| chr3_87030981_87031137 | 0.39 |
Gm8869 |
predicted gene 8869 |
7176 |
0.14 |
| chr7_143498695_143498856 | 0.38 |
Phlda2 |
pleckstrin homology like domain, family A, member 2 |
3766 |
0.14 |
| chr5_73264643_73264803 | 0.38 |
Gm34411 |
predicted gene, 34411 |
1294 |
0.28 |
| chr18_65097598_65097767 | 0.38 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
14628 |
0.23 |
| chr19_46714979_46715130 | 0.38 |
As3mt |
arsenic (+3 oxidation state) methyltransferase |
3082 |
0.19 |
| chr5_53078478_53078847 | 0.38 |
Slc34a2 |
solute carrier family 34 (sodium phosphate), member 2 |
29309 |
0.14 |
| chr4_58546190_58546357 | 0.38 |
Lpar1 |
lysophosphatidic acid receptor 1 |
2128 |
0.33 |
| chr9_63668708_63668859 | 0.38 |
Smad3 |
SMAD family member 3 |
2236 |
0.33 |
| chr7_14425285_14425519 | 0.38 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
11045 |
0.16 |
| chr6_99271808_99271987 | 0.38 |
Foxp1 |
forkhead box P1 |
5365 |
0.31 |
| chr3_37634750_37634916 | 0.38 |
Spry1 |
sprouty RTK signaling antagonist 1 |
5114 |
0.12 |
| chr15_27525366_27525772 | 0.38 |
B230362B09Rik |
RIKEN cDNA B230362B09 gene |
21657 |
0.15 |
| chr1_190156610_190156822 | 0.38 |
Gm28172 |
predicted gene 28172 |
11954 |
0.18 |
| chr5_96458803_96458986 | 0.38 |
Gm33050 |
predicted gene, 33050 |
1151 |
0.56 |
| chr2_44920626_44920948 | 0.37 |
Gtdc1 |
glycosyltransferase-like domain containing 1 |
6362 |
0.29 |
| chr8_126723593_126723744 | 0.37 |
Gm45805 |
predicted gene 45805 |
34666 |
0.19 |
| chr6_128333409_128333573 | 0.37 |
Tulp3 |
tubby-like protein 3 |
7944 |
0.07 |
| chr11_109478088_109478523 | 0.37 |
Slc16a6 |
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
4707 |
0.13 |
| chr18_75891950_75892137 | 0.37 |
Zbtb7c |
zinc finger and BTB domain containing 7C |
71828 |
0.11 |
| chr14_54723804_54723977 | 0.37 |
Cebpe |
CCAAT/enhancer binding protein (C/EBP), epsilon |
11716 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.4 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.4 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.2 | GO:0043465 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.1 | 0.4 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.2 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.2 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.0 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.0 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.2 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0051352 | negative regulation of ligase activity(GO:0051352) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.0 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.3 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.4 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.0 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.0 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.0 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.0 | GO:0046077 | dUDP biosynthetic process(GO:0006227) dTTP biosynthetic process(GO:0006235) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.2 | GO:0052696 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.3 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.2 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0015254 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.0 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |