Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
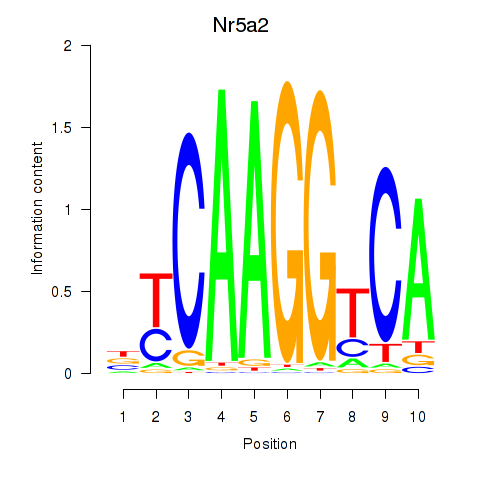
Results for Nr5a2
Z-value: 1.51
Transcription factors associated with Nr5a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr5a2
|
ENSMUSG00000026398.8 | nuclear receptor subfamily 5, group A, member 2 |
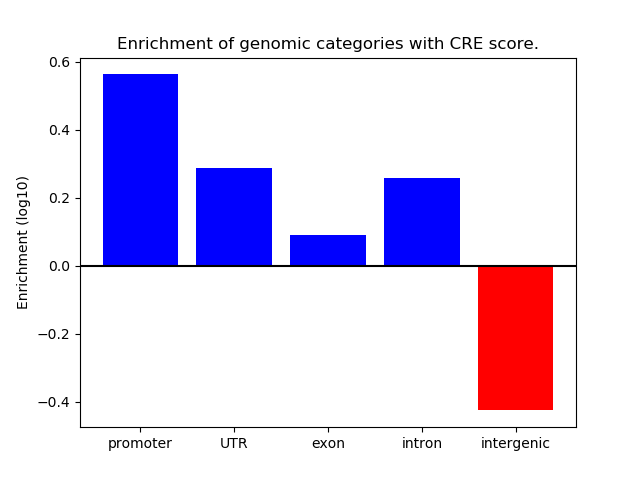
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_136911239_136911410 | Nr5a2 | 29259 | 0.182532 | 0.84 | 3.7e-02 | Click! |
| chr1_136893539_136893740 | Nr5a2 | 46944 | 0.132559 | 0.82 | 4.7e-02 | Click! |
| chr1_136910524_136910904 | Nr5a2 | 29869 | 0.180781 | 0.81 | 5.0e-02 | Click! |
| chr1_136933160_136933350 | Nr5a2 | 7328 | 0.241506 | 0.75 | 8.5e-02 | Click! |
| chr1_136874160_136874323 | Nr5a2 | 66342 | 0.087863 | 0.72 | 1.0e-01 | Click! |
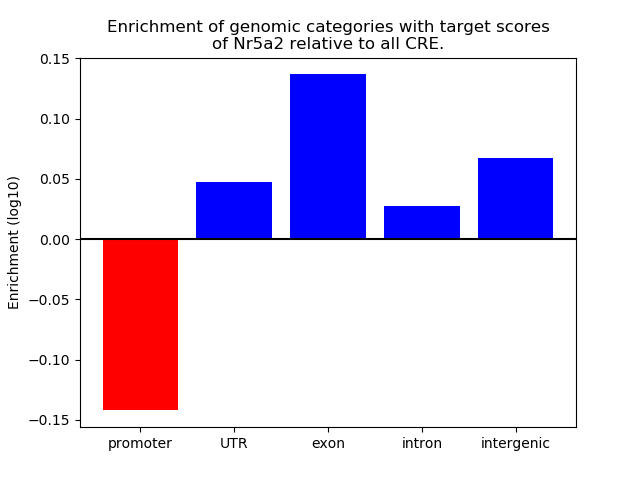
Activity of the Nr5a2 motif across conditions
Conditions sorted by the z-value of the Nr5a2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
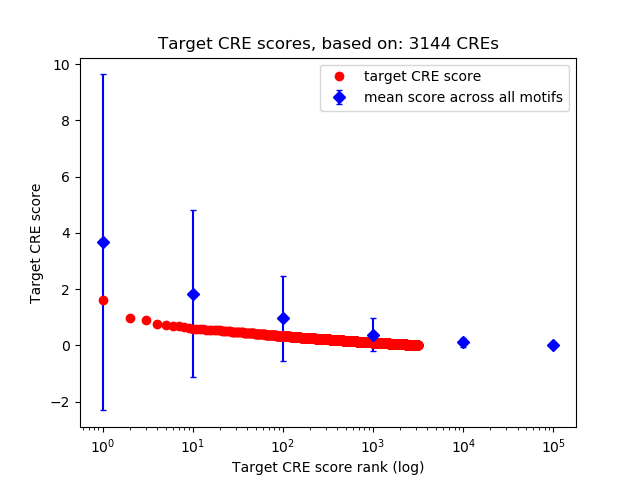
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_182500736_182501185 | 1.61 |
Gm37069 |
predicted gene, 37069 |
6804 |
0.14 |
| chr2_31476173_31476462 | 0.96 |
Ass1 |
argininosuccinate synthetase 1 |
6110 |
0.2 |
| chr10_84418574_84418728 | 0.91 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
21946 |
0.15 |
| chr2_125617617_125617780 | 0.76 |
Cep152 |
centrosomal protein 152 |
7415 |
0.23 |
| chr2_173158545_173158939 | 0.72 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
5660 |
0.18 |
| chr10_24777725_24777876 | 0.70 |
Enpp3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
7205 |
0.23 |
| chr11_7196610_7196788 | 0.69 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1083 |
0.48 |
| chr5_66421556_66421781 | 0.64 |
Gm43792 |
predicted gene 43792 |
10458 |
0.16 |
| chr17_47444308_47444531 | 0.60 |
1700001C19Rik |
RIKEN cDNA 1700001C19 gene |
7043 |
0.12 |
| chr2_34486208_34486368 | 0.59 |
Mapkap1 |
mitogen-activated protein kinase associated protein 1 |
41941 |
0.13 |
| chr13_51095199_51095382 | 0.58 |
Spin1 |
spindlin 1 |
5590 |
0.26 |
| chr6_117297266_117297417 | 0.57 |
Rpl28-ps4 |
ribosomal protein L28, pseudogene 4 |
83275 |
0.08 |
| chr3_18626956_18627107 | 0.57 |
Gm42944 |
predicted gene 42944 |
4869 |
0.23 |
| chr8_105044924_105045225 | 0.55 |
Gm45725 |
predicted gene 45725 |
92 |
0.94 |
| chr8_10935282_10935610 | 0.55 |
Gm45042 |
predicted gene 45042 |
5559 |
0.11 |
| chr12_52442391_52442588 | 0.55 |
Gm47431 |
predicted gene, 47431 |
5636 |
0.23 |
| chr7_73587321_73587472 | 0.54 |
Gm44734 |
predicted gene 44734 |
21552 |
0.09 |
| chr10_8229677_8229834 | 0.54 |
Gm30906 |
predicted gene, 30906 |
53624 |
0.15 |
| chr5_134456918_134457072 | 0.53 |
Gtf2ird1 |
general transcription factor II I repeat domain-containing 1 |
279 |
0.77 |
| chr10_21447624_21447924 | 0.53 |
Gm48386 |
predicted gene, 48386 |
2138 |
0.25 |
| chr18_3415919_3416074 | 0.52 |
Cul2 |
cullin 2 |
17888 |
0.15 |
| chr8_33798912_33799096 | 0.51 |
Rbpms |
RNA binding protein gene with multiple splicing |
1790 |
0.29 |
| chr19_30150086_30150237 | 0.50 |
Gldc |
glycine decarboxylase |
4930 |
0.21 |
| chr2_69466274_69466471 | 0.50 |
Lrp2 |
low density lipoprotein receptor-related protein 2 |
42773 |
0.15 |
| chr17_12392360_12392620 | 0.49 |
Plg |
plasminogen |
13831 |
0.16 |
| chr6_72571781_72571932 | 0.49 |
Capg |
capping protein (actin filament), gelsolin-like |
15877 |
0.09 |
| chr14_25516055_25516210 | 0.48 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
13477 |
0.15 |
| chr17_12411169_12411345 | 0.48 |
Plg |
plasminogen |
32598 |
0.13 |
| chr10_96235051_96235242 | 0.48 |
4930459C07Rik |
RIKEN cDNA 4930459C07 gene |
8939 |
0.2 |
| chr8_119443050_119443280 | 0.47 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
3554 |
0.18 |
| chr1_51877379_51877530 | 0.47 |
Gm28323 |
predicted gene 28323 |
2154 |
0.26 |
| chr9_48767602_48767753 | 0.46 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
68268 |
0.11 |
| chr6_141432376_141432556 | 0.46 |
Gm43958 |
predicted gene, 43958 |
9894 |
0.26 |
| chr1_178219599_178219928 | 0.46 |
Desi2 |
desumoylating isopeptidase 2 |
31916 |
0.13 |
| chr9_20741226_20741380 | 0.46 |
Olfm2 |
olfactomedin 2 |
5046 |
0.17 |
| chr1_51774175_51774474 | 0.46 |
Myo1b |
myosin IB |
2112 |
0.33 |
| chr5_150227352_150227751 | 0.45 |
Gm36378 |
predicted gene, 36378 |
16312 |
0.19 |
| chr9_43622435_43622586 | 0.45 |
Gm29909 |
predicted gene, 29909 |
14326 |
0.17 |
| chr2_163595110_163595261 | 0.45 |
Ttpal |
tocopherol (alpha) transfer protein-like |
7129 |
0.15 |
| chr7_112294455_112294629 | 0.45 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
21487 |
0.25 |
| chr4_145016736_145017043 | 0.44 |
Vps13d |
vacuolar protein sorting 13D |
32928 |
0.18 |
| chr19_45310205_45310467 | 0.44 |
Gm28578 |
predicted gene 28578 |
754 |
0.62 |
| chr15_11394606_11394765 | 0.43 |
Tars |
threonyl-tRNA synthetase |
2415 |
0.37 |
| chr13_43253808_43253976 | 0.43 |
Gfod1 |
glucose-fructose oxidoreductase domain containing 1 |
49513 |
0.13 |
| chr2_173159655_173159816 | 0.43 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
6653 |
0.18 |
| chr5_125325436_125325605 | 0.43 |
Scarb1 |
scavenger receptor class B, member 1 |
747 |
0.57 |
| chr2_153580884_153581050 | 0.42 |
Nol4l |
nucleolar protein 4-like |
50996 |
0.1 |
| chr2_172472866_172473017 | 0.42 |
Fam209 |
family with sequence similarity 209 |
421 |
0.78 |
| chr4_19523139_19523419 | 0.42 |
Cpne3 |
copine III |
20399 |
0.22 |
| chr7_28732426_28732636 | 0.42 |
Fbxo17 |
F-box protein 17 |
31 |
0.95 |
| chr15_102179533_102179684 | 0.41 |
Csad |
cysteine sulfinic acid decarboxylase |
253 |
0.85 |
| chr14_73045910_73046229 | 0.41 |
Cysltr2 |
cysteinyl leukotriene receptor 2 |
3003 |
0.3 |
| chr14_71963090_71963264 | 0.41 |
4930434J06Rik |
RIKEN cDNA 4930434J06 gene |
50086 |
0.18 |
| chr11_98301768_98301944 | 0.41 |
Gm20644 |
predicted gene 20644 |
16474 |
0.09 |
| chr1_156184131_156184502 | 0.41 |
Fam163a |
family with sequence similarity 163, member A |
20710 |
0.16 |
| chr2_129337674_129337837 | 0.40 |
Gm25703 |
predicted gene, 25703 |
22276 |
0.09 |
| chr10_5130565_5130739 | 0.40 |
Syne1 |
spectrin repeat containing, nuclear envelope 1 |
11876 |
0.22 |
| chr5_114558713_114558889 | 0.40 |
Gm13790 |
predicted gene 13790 |
7512 |
0.16 |
| chr17_12931620_12931776 | 0.40 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
4016 |
0.09 |
| chr4_141276769_141276920 | 0.39 |
Gm13056 |
predicted gene 13056 |
1595 |
0.24 |
| chr10_77008035_77008232 | 0.39 |
Gm35721 |
predicted gene, 35721 |
436 |
0.75 |
| chr16_29943589_29943740 | 0.39 |
Gm26569 |
predicted gene, 26569 |
2852 |
0.26 |
| chr10_59784742_59784904 | 0.39 |
Gm17059 |
predicted gene 17059 |
15431 |
0.14 |
| chr3_127103964_127104222 | 0.38 |
Ank2 |
ankyrin 2, brain |
20769 |
0.14 |
| chr9_26413705_26413886 | 0.38 |
Gm48373 |
predicted gene, 48373 |
27493 |
0.21 |
| chr16_31567276_31567455 | 0.38 |
Gm34256 |
predicted gene, 34256 |
25855 |
0.14 |
| chr14_59443633_59443793 | 0.38 |
Cab39l |
calcium binding protein 39-like |
2732 |
0.21 |
| chr14_20148641_20148813 | 0.38 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
3433 |
0.19 |
| chr2_41790211_41790364 | 0.38 |
Lrp1b |
low density lipoprotein-related protein 1B |
1209 |
0.66 |
| chr1_86998138_86998337 | 0.38 |
Gm37017 |
predicted gene, 37017 |
9967 |
0.13 |
| chr10_66914376_66914558 | 0.38 |
1110002J07Rik |
RIKEN cDNA 1110002J07 gene |
3272 |
0.2 |
| chr3_117862558_117862719 | 0.37 |
Snx7 |
sorting nexin 7 |
6183 |
0.21 |
| chr8_122274769_122274938 | 0.37 |
Zfp469 |
zinc finger protein 469 |
5284 |
0.17 |
| chr2_30759973_30760153 | 0.37 |
1700001O22Rik |
RIKEN cDNA 1700001O22 gene |
36601 |
0.09 |
| chr5_92206260_92206413 | 0.37 |
U90926 |
cDNA sequence U90926 |
6014 |
0.11 |
| chr9_65334133_65334359 | 0.37 |
Gm39363 |
predicted gene, 39363 |
1726 |
0.18 |
| chr17_31861640_31861807 | 0.37 |
Sik1 |
salt inducible kinase 1 |
5919 |
0.17 |
| chr14_65721753_65721925 | 0.36 |
Scara5 |
scavenger receptor class A, member 5 |
45362 |
0.14 |
| chr1_102517589_102517767 | 0.36 |
Gm20281 |
predicted gene, 20281 |
58756 |
0.14 |
| chr2_59979903_59980091 | 0.35 |
Gm13574 |
predicted gene 13574 |
12347 |
0.19 |
| chr5_135879620_135879793 | 0.35 |
Gm22450 |
predicted gene, 22450 |
4049 |
0.12 |
| chr18_20980054_20980205 | 0.35 |
Rnf125 |
ring finger protein 125 |
18657 |
0.18 |
| chr17_31640162_31640374 | 0.35 |
Cbs |
cystathionine beta-synthase |
3030 |
0.13 |
| chr11_52220968_52221143 | 0.35 |
Olfr1371 |
olfactory receptor 1371 |
7068 |
0.12 |
| chr12_85112663_85112863 | 0.35 |
Dlst |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
1870 |
0.19 |
| chr4_40849235_40849401 | 0.34 |
Mir5123 |
microRNA 5123 |
820 |
0.3 |
| chr8_119887445_119887613 | 0.34 |
Usp10 |
ubiquitin specific peptidase 10 |
22831 |
0.13 |
| chr17_29061153_29061388 | 0.34 |
Gm41556 |
predicted gene, 41556 |
3041 |
0.12 |
| chr18_20989752_20989946 | 0.34 |
Rnf138 |
ring finger protein 138 |
11492 |
0.2 |
| chr3_86018925_86019076 | 0.34 |
Prss48 |
protease, serine 48 |
16509 |
0.12 |
| chrX_77607586_77607742 | 0.34 |
Gm23121 |
predicted gene, 23121 |
28177 |
0.18 |
| chr9_48650755_48650912 | 0.34 |
Nnmt |
nicotinamide N-methyltransferase |
45680 |
0.16 |
| chr10_77587179_77587612 | 0.33 |
Pttg1ip |
pituitary tumor-transforming 1 interacting protein |
1999 |
0.18 |
| chr6_113011965_113012136 | 0.33 |
Gm8083 |
predicted gene 8083 |
19928 |
0.09 |
| chr2_125617422_125617574 | 0.33 |
Cep152 |
centrosomal protein 152 |
7615 |
0.22 |
| chr4_132476053_132476227 | 0.33 |
Med18 |
mediator complex subunit 18 |
12219 |
0.1 |
| chr4_46527475_46527653 | 0.33 |
Trim14 |
tripartite motif-containing 14 |
6850 |
0.14 |
| chr18_65058969_65059130 | 0.33 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
8059 |
0.24 |
| chr9_110703228_110703412 | 0.33 |
Ccdc12 |
coiled-coil domain containing 12 |
6574 |
0.12 |
| chr1_51996762_51997232 | 0.33 |
Stat4 |
signal transducer and activator of transcription 4 |
9849 |
0.17 |
| chr7_44898673_44898846 | 0.33 |
Fuz |
fuzzy planar cell polarity protein |
492 |
0.42 |
| chr1_180179778_180180226 | 0.33 |
Coq8a |
coenzyme Q8A |
1080 |
0.44 |
| chr4_132960785_132960960 | 0.33 |
Fgr |
FGR proto-oncogene, Src family tyrosine kinase |
13223 |
0.14 |
| chrX_169904110_169904261 | 0.33 |
Mid1 |
midline 1 |
22766 |
0.2 |
| chrX_12377036_12377378 | 0.32 |
Gm14635 |
predicted gene 14635 |
22211 |
0.24 |
| chr8_75092030_75092181 | 0.32 |
Hmox1 |
heme oxygenase 1 |
1516 |
0.3 |
| chr14_68809096_68809247 | 0.32 |
Gm47256 |
predicted gene, 47256 |
65664 |
0.11 |
| chr5_151103422_151103608 | 0.32 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
5220 |
0.28 |
| chr9_115425112_115425263 | 0.32 |
Gm5921 |
predicted gene 5921 |
13394 |
0.13 |
| chr8_126823704_126823868 | 0.32 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
15447 |
0.2 |
| chr6_115838733_115838884 | 0.32 |
Efcab12 |
EF-hand calcium binding domain 12 |
396 |
0.72 |
| chr5_140167245_140167441 | 0.32 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
7316 |
0.17 |
| chr2_156433040_156433199 | 0.32 |
Epb41l1 |
erythrocyte membrane protein band 4.1 like 1 |
12007 |
0.09 |
| chr17_70753482_70753646 | 0.32 |
5031415H12Rik |
RIKEN cDNA 5031415H12 gene |
2018 |
0.31 |
| chr19_29520832_29521011 | 0.32 |
A930007I19Rik |
RIKEN cDNA A930007I19 gene |
49 |
0.96 |
| chr1_190117320_190117686 | 0.32 |
Gm28172 |
predicted gene 28172 |
51167 |
0.13 |
| chr16_93918744_93918905 | 0.32 |
Cldn14 |
claudin 14 |
10743 |
0.14 |
| chr1_91822004_91822314 | 0.31 |
Gm29101 |
predicted gene 29101 |
356 |
0.87 |
| chr6_72212494_72212706 | 0.31 |
Atoh8 |
atonal bHLH transcription factor 8 |
21937 |
0.14 |
| chr16_13257357_13257584 | 0.31 |
Mrtfb |
myocardin related transcription factor B |
964 |
0.67 |
| chr12_8829085_8829244 | 0.31 |
9930038B18Rik |
RIKEN cDNA 9930038B18 gene |
48218 |
0.11 |
| chr5_120484543_120484824 | 0.31 |
Gm15690 |
predicted gene 15690 |
3780 |
0.13 |
| chr3_131473184_131473335 | 0.31 |
Sgms2 |
sphingomyelin synthase 2 |
17220 |
0.23 |
| chr2_34828383_34828580 | 0.31 |
Fbxw2 |
F-box and WD-40 domain protein 2 |
2170 |
0.17 |
| chr5_66232678_66232831 | 0.31 |
Gm3822 |
predicted gene 3822 |
18007 |
0.11 |
| chr12_69366525_69366676 | 0.31 |
Gm18113 |
predicted gene, 18113 |
3423 |
0.12 |
| chr8_83964735_83964914 | 0.30 |
Asf1b |
anti-silencing function 1B histone chaperone |
223 |
0.83 |
| chr3_36480639_36480833 | 0.30 |
1810062G17Rik |
RIKEN cDNA 1810062G17 gene |
4799 |
0.13 |
| chr19_20605270_20605421 | 0.30 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
3384 |
0.27 |
| chr12_114378675_114378838 | 0.30 |
Ighv1-1 |
immunoglobulin heavy variable V1-1 |
26761 |
0.1 |
| chr3_63315218_63315408 | 0.30 |
Mme |
membrane metallo endopeptidase |
15153 |
0.26 |
| chr6_124905504_124905662 | 0.30 |
Lag3 |
lymphocyte-activation gene 3 |
4468 |
0.09 |
| chr19_38370339_38370560 | 0.30 |
Gm50155 |
predicted gene, 50155 |
15065 |
0.13 |
| chr16_91931256_91931425 | 0.30 |
Atp5o |
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
66 |
0.54 |
| chr16_10524923_10525085 | 0.30 |
Dexi |
dexamethasone-induced transcript |
6805 |
0.18 |
| chr11_95744707_95744876 | 0.29 |
Zfp652 |
zinc finger protein 652 |
4209 |
0.15 |
| chr2_77508328_77508502 | 0.29 |
Zfp385b |
zinc finger protein 385B |
11120 |
0.25 |
| chr8_3390517_3390679 | 0.29 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
2408 |
0.27 |
| chr11_100771131_100771291 | 0.29 |
Ghdc |
GH3 domain containing |
254 |
0.85 |
| chr7_51899481_51899655 | 0.29 |
Gas2 |
growth arrest specific 2 |
11650 |
0.17 |
| chr5_104374044_104374195 | 0.29 |
Mepe |
matrix extracellular phosphoglycoprotein with ASARM motif (bone) |
48790 |
0.1 |
| chr14_68304879_68305239 | 0.29 |
Gm47212 |
predicted gene, 47212 |
86806 |
0.08 |
| chr7_30421568_30421733 | 0.29 |
Nfkbid |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
82 |
0.91 |
| chr13_37824271_37824422 | 0.29 |
Rreb1 |
ras responsive element binding protein 1 |
1629 |
0.37 |
| chr4_107920456_107920610 | 0.29 |
Cpt2 |
carnitine palmitoyltransferase 2 |
2914 |
0.18 |
| chr4_103115043_103115199 | 0.29 |
Mier1 |
MEIR1 treanscription regulator |
151 |
0.84 |
| chr6_93770976_93771287 | 0.29 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
21408 |
0.21 |
| chr7_143296870_143297021 | 0.28 |
Kcnq1ot1 |
KCNQ1 overlapping transcript 1 |
396 |
0.65 |
| chr15_102177432_102177780 | 0.28 |
Csad |
cysteine sulfinic acid decarboxylase |
1413 |
0.26 |
| chr13_37953178_37953339 | 0.28 |
Rreb1 |
ras responsive element binding protein 1 |
6242 |
0.21 |
| chr7_25448767_25449204 | 0.28 |
Gm15495 |
predicted gene 15495 |
5522 |
0.11 |
| chr4_96583762_96583984 | 0.28 |
Cyp2j9 |
cytochrome P450, family 2, subfamily j, polypeptide 9 |
7705 |
0.22 |
| chrX_10556988_10557139 | 0.28 |
Gm25885 |
predicted gene, 25885 |
47015 |
0.14 |
| chr9_70069929_70070289 | 0.28 |
Gm47233 |
predicted gene, 47233 |
242 |
0.89 |
| chr19_44128120_44128295 | 0.28 |
Cwf19l1 |
CWF19-like 1, cell cycle control (S. pombe) |
5291 |
0.13 |
| chr9_61911528_61911679 | 0.28 |
Rplp1 |
ribosomal protein, large, P1 |
2939 |
0.26 |
| chr12_110021918_110022099 | 0.28 |
Gm34667 |
predicted gene, 34667 |
1865 |
0.26 |
| chr6_30564201_30564366 | 0.28 |
Cpa4 |
carboxypeptidase A4 |
4086 |
0.15 |
| chr18_46649890_46650075 | 0.28 |
Gm3734 |
predicted gene 3734 |
19175 |
0.14 |
| chr4_136173095_136173459 | 0.28 |
E2f2 |
E2F transcription factor 2 |
883 |
0.51 |
| chr18_61424158_61424549 | 0.28 |
Gm8755 |
predicted gene 8755 |
9848 |
0.12 |
| chr4_63558749_63559072 | 0.28 |
Tmem268 |
transmembrane protein 268 |
54 |
0.96 |
| chr12_35044157_35044343 | 0.28 |
Snx13 |
sorting nexin 13 |
2936 |
0.27 |
| chr1_133113123_133113283 | 0.28 |
Ppp1r15b |
protein phosphatase 1, regulatory subunit 15B |
17940 |
0.13 |
| chr3_88833523_88833680 | 0.28 |
1500004A13Rik |
RIKEN cDNA 1500004A13 gene |
1054 |
0.32 |
| chr9_80629501_80629676 | 0.27 |
Gm48387 |
predicted gene, 48387 |
28491 |
0.22 |
| chr8_70170192_70170368 | 0.27 |
Tmem161a |
transmembrane protein 161A |
2076 |
0.16 |
| chr5_66079062_66079213 | 0.27 |
Rbm47 |
RNA binding motif protein 47 |
1847 |
0.24 |
| chr11_55062992_55063153 | 0.27 |
Ccdc69 |
coiled-coil domain containing 69 |
15035 |
0.13 |
| chr6_128523521_128524414 | 0.27 |
Pzp |
PZP, alpha-2-macroglobulin like |
2736 |
0.13 |
| chr10_21686247_21686409 | 0.27 |
Gm5420 |
predicted gene 5420 |
83 |
0.98 |
| chr17_25727890_25728059 | 0.27 |
Rpusd1 |
RNA pseudouridylate synthase domain containing 1 |
242 |
0.65 |
| chr11_88718878_88719286 | 0.27 |
C030037D09Rik |
RIKEN cDNA C030037D09 gene |
278 |
0.72 |
| chr5_135167079_135167237 | 0.27 |
Bcl7b |
B cell CLL/lymphoma 7B |
1125 |
0.34 |
| chr3_103025386_103025549 | 0.27 |
Csde1 |
cold shock domain containing E1, RNA binding |
3535 |
0.16 |
| chr17_57058382_57058541 | 0.27 |
Crb3 |
crumbs family member 3 |
644 |
0.4 |
| chr15_27822193_27822368 | 0.27 |
Trio |
triple functional domain (PTPRF interacting) |
1503 |
0.44 |
| chr15_103085855_103086027 | 0.27 |
Gm28876 |
predicted gene 28876 |
16170 |
0.09 |
| chr19_20450928_20451527 | 0.27 |
C730002L08Rik |
RIKEN cDNA C730002L08 gene |
26445 |
0.16 |
| chr6_50773105_50773295 | 0.27 |
C530044C16Rik |
RIKEN cDNA C530044C16 gene |
2915 |
0.22 |
| chr16_46098109_46098276 | 0.27 |
Gm50487 |
predicted gene, 50487 |
11717 |
0.18 |
| chr12_17445944_17446108 | 0.27 |
Gm36752 |
predicted gene, 36752 |
9408 |
0.2 |
| chr18_20938674_20939098 | 0.27 |
Rnf125 |
ring finger protein 125 |
5739 |
0.23 |
| chr1_177664080_177664231 | 0.27 |
2310043L19Rik |
RIKEN cDNA 2310043L19 gene |
21212 |
0.16 |
| chr5_151246921_151247115 | 0.26 |
5430435K18Rik |
RIKEN cDNA 5430435K18 gene |
5287 |
0.21 |
| chr19_3358921_3359072 | 0.26 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
7334 |
0.14 |
| chr8_124923216_124923378 | 0.26 |
Sprtn |
SprT-like N-terminal domain |
21482 |
0.11 |
| chr4_141115485_141115659 | 0.26 |
4921514A10Rik |
RIKEN cDNA 4921514A10 gene |
88 |
0.95 |
| chr6_6997102_6997263 | 0.26 |
Sdhaf3 |
succinate dehydrogenase complex assembly factor 3 |
41175 |
0.13 |
| chr2_144014015_144014533 | 0.26 |
Rrbp1 |
ribosome binding protein 1 |
3011 |
0.24 |
| chr8_126919454_126919605 | 0.26 |
Gm31718 |
predicted gene, 31718 |
11314 |
0.16 |
| chr2_109694324_109694484 | 0.26 |
Bdnf |
brain derived neurotrophic factor |
5 |
0.98 |
| chr7_70800023_70800227 | 0.26 |
Gm24880 |
predicted gene, 24880 |
47709 |
0.15 |
| chr4_44990961_44991112 | 0.26 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
9536 |
0.11 |
| chr14_66004934_66005102 | 0.26 |
Gulo |
gulonolactone (L-) oxidase |
4189 |
0.18 |
| chr17_79919609_79919795 | 0.26 |
Gm6552 |
predicted gene 6552 |
14870 |
0.16 |
| chr14_64445994_64446184 | 0.26 |
Msra |
methionine sulfoxide reductase A |
5172 |
0.28 |
| chr11_97361922_97362073 | 0.26 |
Socs7 |
suppressor of cytokine signaling 7 |
438 |
0.76 |
| chr5_150603699_150604014 | 0.26 |
Gm43597 |
predicted gene 43597 |
3027 |
0.12 |
| chr17_87102953_87103253 | 0.26 |
Socs5 |
suppressor of cytokine signaling 5 |
4576 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.3 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.2 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.2 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.3 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.0 | 0.2 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.4 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.2 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0052173 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:0061623 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.0 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.0 | 0.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0051794 | regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.0 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.0 | GO:0046122 | dGTP metabolic process(GO:0046070) purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.0 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0047419 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |