Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
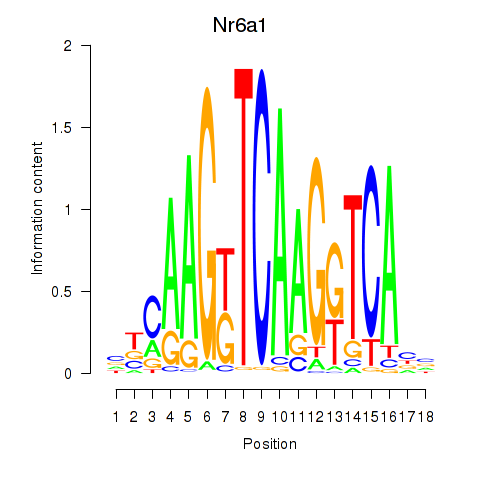
Results for Nr6a1
Z-value: 2.09
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSMUSG00000063972.7 | nuclear receptor subfamily 6, group A, member 1 |
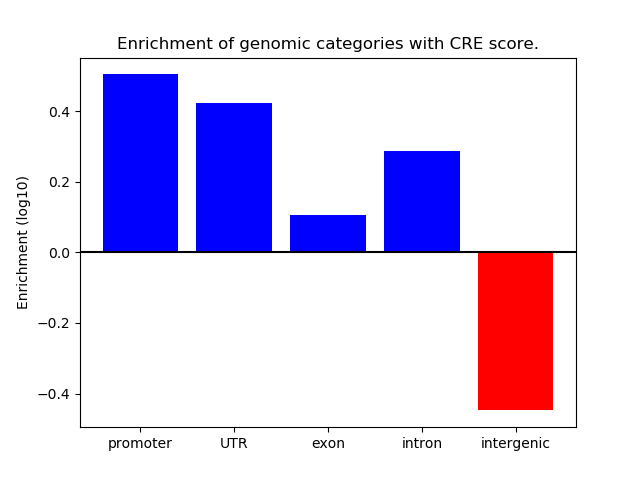
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_38782536_38782710 | Nr6a1 | 1846 | 0.222365 | -0.89 | 1.9e-02 | Click! |
| chr2_38785061_38785421 | Nr6a1 | 130 | 0.935081 | -0.79 | 6.3e-02 | Click! |
| chr2_38926124_38926288 | Nr6a1 | 11 | 0.958036 | 0.76 | 8.2e-02 | Click! |
| chr2_38786516_38786686 | Nr6a1 | 1490 | 0.273857 | -0.57 | 2.4e-01 | Click! |
| chr2_38925438_38925645 | Nr6a1 | 676 | 0.543889 | 0.57 | 2.4e-01 | Click! |
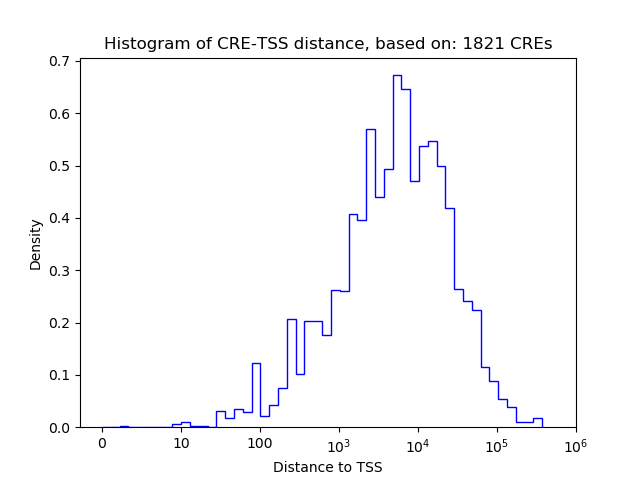
Activity of the Nr6a1 motif across conditions
Conditions sorted by the z-value of the Nr6a1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
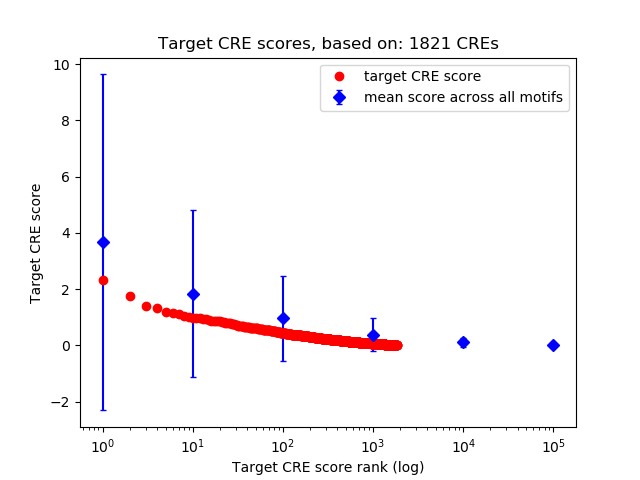
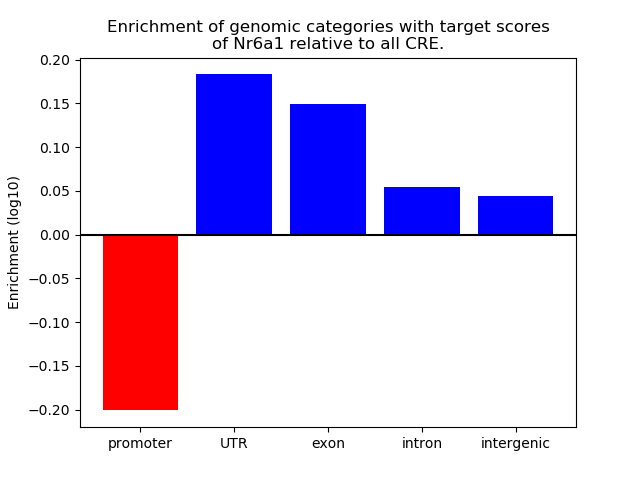
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_93618635_93618791 | 2.32 |
Gm15622 |
predicted gene 15622 |
6669 |
0.17 |
| chr1_182500736_182501185 | 1.74 |
Gm37069 |
predicted gene, 37069 |
6804 |
0.14 |
| chr10_8229677_8229834 | 1.39 |
Gm30906 |
predicted gene, 30906 |
53624 |
0.15 |
| chr18_20987436_20987590 | 1.34 |
Rnf138 |
ring finger protein 138 |
13828 |
0.19 |
| chr2_172472866_172473017 | 1.19 |
Fam209 |
family with sequence similarity 209 |
421 |
0.78 |
| chr16_32685796_32686168 | 1.15 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
5168 |
0.16 |
| chr17_12411169_12411345 | 1.11 |
Plg |
plasminogen |
32598 |
0.13 |
| chr1_72854037_72854202 | 1.05 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
5420 |
0.23 |
| chr17_31861640_31861807 | 1.02 |
Sik1 |
salt inducible kinase 1 |
5919 |
0.17 |
| chr15_102179533_102179684 | 0.98 |
Csad |
cysteine sulfinic acid decarboxylase |
253 |
0.85 |
| chr3_131473184_131473335 | 0.97 |
Sgms2 |
sphingomyelin synthase 2 |
17220 |
0.23 |
| chr2_164054230_164054392 | 0.95 |
Pabpc1l |
poly(A) binding protein, cytoplasmic 1-like |
6301 |
0.14 |
| chr6_97901721_97901885 | 0.93 |
Gm15531 |
predicted gene 15531 |
11264 |
0.23 |
| chr10_21916464_21916630 | 0.93 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
11923 |
0.19 |
| chr4_44992225_44992598 | 0.91 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
10911 |
0.1 |
| chr2_69406819_69407032 | 0.87 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
26480 |
0.18 |
| chr7_112294455_112294629 | 0.86 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
21487 |
0.25 |
| chr8_84068264_84068441 | 0.86 |
C330011M18Rik |
RIKEN cDNA C330011M18 gene |
1065 |
0.23 |
| chr6_4820679_4820851 | 0.85 |
Gm20714 |
predicted gene 20714 |
4436 |
0.18 |
| chr13_60709722_60709957 | 0.85 |
Dapk1 |
death associated protein kinase 1 |
13426 |
0.17 |
| chr1_40132911_40133091 | 0.83 |
Gm37347 |
predicted gene, 37347 |
44490 |
0.11 |
| chr9_121913349_121913521 | 0.82 |
Cyp8b1 |
cytochrome P450, family 8, subfamily b, polypeptide 1 |
2870 |
0.12 |
| chr2_173974366_173974517 | 0.80 |
Atp5k-ps2 |
ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E, pseudogene 2 |
10250 |
0.2 |
| chr13_91762116_91762414 | 0.78 |
Gm27656 |
predicted gene, 27656 |
10159 |
0.15 |
| chrX_77607586_77607742 | 0.78 |
Gm23121 |
predicted gene, 23121 |
28177 |
0.18 |
| chr19_47385384_47385539 | 0.78 |
Sh3pxd2a |
SH3 and PX domains 2A |
24898 |
0.18 |
| chr13_64242805_64242976 | 0.77 |
Cdc14b |
CDC14 cell division cycle 14B |
5752 |
0.11 |
| chr9_20741226_20741380 | 0.76 |
Olfm2 |
olfactomedin 2 |
5046 |
0.17 |
| chr17_33807244_33807514 | 0.75 |
Kank3 |
KN motif and ankyrin repeat domains 3 |
3140 |
0.1 |
| chr5_135167079_135167237 | 0.72 |
Bcl7b |
B cell CLL/lymphoma 7B |
1125 |
0.34 |
| chr17_33932704_33933052 | 0.71 |
Rgl2 |
ral guanine nucleotide dissociation stimulator-like 2 |
747 |
0.27 |
| chr1_51877379_51877530 | 0.70 |
Gm28323 |
predicted gene 28323 |
2154 |
0.26 |
| chr17_84037207_84037358 | 0.70 |
Gm49967 |
predicted gene, 49967 |
392 |
0.78 |
| chr13_13558705_13559076 | 0.69 |
Lyst |
lysosomal trafficking regulator |
31507 |
0.14 |
| chr3_96402109_96402496 | 0.69 |
Gm26654 |
predicted gene, 26654 |
79 |
0.72 |
| chr19_47276101_47276391 | 0.67 |
Mir6995 |
microRNA 6995 |
2657 |
0.21 |
| chr4_98108774_98108954 | 0.66 |
Gm12691 |
predicted gene 12691 |
37735 |
0.2 |
| chr9_106194059_106194232 | 0.65 |
Ppm1m |
protein phosphatase 1M |
2743 |
0.13 |
| chr5_8990837_8991059 | 0.65 |
Crot |
carnitine O-octanoyltransferase |
6094 |
0.13 |
| chr15_11394606_11394765 | 0.65 |
Tars |
threonyl-tRNA synthetase |
2415 |
0.37 |
| chr15_76324787_76324950 | 0.64 |
Exosc4 |
exosome component 4 |
2529 |
0.09 |
| chr14_31653594_31653749 | 0.64 |
Hacl1 |
2-hydroxyacyl-CoA lyase 1 |
12385 |
0.14 |
| chr4_132974220_132974503 | 0.62 |
Fgr |
FGR proto-oncogene, Src family tyrosine kinase |
259 |
0.9 |
| chr12_85112663_85112863 | 0.62 |
Dlst |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
1870 |
0.19 |
| chr8_10935282_10935610 | 0.62 |
Gm45042 |
predicted gene 45042 |
5559 |
0.11 |
| chr2_53000554_53000705 | 0.62 |
Fmnl2 |
formin-like 2 |
142761 |
0.04 |
| chr5_120501180_120501331 | 0.62 |
Plbd2 |
phospholipase B domain containing 2 |
2346 |
0.17 |
| chr6_124872495_124872656 | 0.61 |
Cd4 |
CD4 antigen |
2086 |
0.13 |
| chr6_87759992_87760347 | 0.61 |
Gm5879 |
predicted gene 5879 |
6035 |
0.09 |
| chr2_44142320_44142937 | 0.61 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
77304 |
0.11 |
| chr11_100771131_100771291 | 0.61 |
Ghdc |
GH3 domain containing |
254 |
0.85 |
| chr2_104736695_104736943 | 0.61 |
Depdc7 |
DEP domain containing 7 |
6059 |
0.16 |
| chr2_173633527_173633866 | 0.59 |
Ppp4r1l-ps |
protein phosphatase 4, regulatory subunit 1-like, pseudogene |
4146 |
0.21 |
| chr13_75793210_75793372 | 0.58 |
Gm8288 |
predicted gene 8288 |
9415 |
0.14 |
| chr6_124905504_124905662 | 0.58 |
Lag3 |
lymphocyte-activation gene 3 |
4468 |
0.09 |
| chr1_150410095_150410265 | 0.58 |
Tpr |
translocated promoter region, nuclear basket protein |
1817 |
0.32 |
| chr1_105822945_105823096 | 0.58 |
Tnfrsf11a |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
3150 |
0.26 |
| chr9_85768264_85768415 | 0.57 |
Gm22830 |
predicted gene, 22830 |
4141 |
0.16 |
| chr17_31640162_31640374 | 0.56 |
Cbs |
cystathionine beta-synthase |
3030 |
0.13 |
| chr18_65058969_65059130 | 0.56 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
8059 |
0.24 |
| chr8_44708968_44709119 | 0.56 |
Gm26089 |
predicted gene, 26089 |
107210 |
0.07 |
| chr14_73136010_73136299 | 0.55 |
Rcbtb2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
2614 |
0.29 |
| chr8_124343978_124344300 | 0.55 |
Gm15775 |
predicted gene 15775 |
12344 |
0.14 |
| chr5_118291975_118292126 | 0.55 |
Gm25076 |
predicted gene, 25076 |
25601 |
0.16 |
| chr2_155279279_155279430 | 0.54 |
Map1lc3a |
microtubule-associated protein 1 light chain 3 alpha |
2465 |
0.24 |
| chr6_72212494_72212706 | 0.54 |
Atoh8 |
atonal bHLH transcription factor 8 |
21937 |
0.14 |
| chr5_32137626_32137972 | 0.53 |
Fosl2 |
fos-like antigen 2 |
1626 |
0.31 |
| chr7_63916741_63916912 | 0.53 |
E030018B13Rik |
RIKEN cDNA E030018B13 gene |
31 |
0.97 |
| chr15_81181427_81181589 | 0.53 |
Mrtfa |
myocardin related transcription factor A |
9014 |
0.14 |
| chr3_14769683_14769846 | 0.53 |
Car1 |
carbonic anhydrase 1 |
8695 |
0.19 |
| chr11_81531658_81531818 | 0.53 |
1700071K01Rik |
RIKEN cDNA 1700071K01 gene |
41735 |
0.18 |
| chr2_126584357_126584536 | 0.53 |
Hdc |
histidine decarboxylase |
13815 |
0.16 |
| chr11_75318270_75318428 | 0.53 |
Gm47300 |
predicted gene, 47300 |
259 |
0.89 |
| chr5_123349021_123349175 | 0.52 |
Bcl7a |
B cell CLL/lymphoma 7A |
4598 |
0.1 |
| chr3_115822649_115822812 | 0.51 |
Dph5 |
diphthamide biosynthesis 5 |
65107 |
0.08 |
| chr3_108070638_108070797 | 0.51 |
Ampd2 |
adenosine monophosphate deaminase 2 |
4753 |
0.09 |
| chr8_107483340_107483510 | 0.51 |
Wwp2 |
WW domain containing E3 ubiquitin protein ligase 2 |
93 |
0.97 |
| chr16_16456437_16456606 | 0.51 |
Fgd4 |
FYVE, RhoGEF and PH domain containing 4 |
47701 |
0.11 |
| chr5_151103422_151103608 | 0.51 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
5220 |
0.28 |
| chr8_126772922_126773091 | 0.50 |
Gm45805 |
predicted gene 45805 |
14672 |
0.23 |
| chr9_108083457_108083608 | 0.49 |
Rnf123 |
ring finger protein 123 |
186 |
0.77 |
| chr6_140748785_140748936 | 0.49 |
Rpl38-ps2 |
ribosomal protein L38, pseudogene 2 |
1688 |
0.38 |
| chr13_93622998_93623302 | 0.49 |
Gm15622 |
predicted gene 15622 |
2232 |
0.26 |
| chr12_113071949_113072118 | 0.48 |
Pacs2 |
phosphofurin acidic cluster sorting protein 2 |
1361 |
0.26 |
| chr3_91463817_91463984 | 0.48 |
S100a7l2 |
S100 calcium binding protein A7 like 2 |
373097 |
0.01 |
| chr19_16479183_16479334 | 0.47 |
Gm8222 |
predicted gene 8222 |
11806 |
0.19 |
| chr14_70530138_70530323 | 0.47 |
Lgi3 |
leucine-rich repeat LGI family, member 3 |
455 |
0.68 |
| chr16_91796689_91796840 | 0.47 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
7858 |
0.19 |
| chr9_36794772_36794937 | 0.47 |
Ei24 |
etoposide induced 2.4 mRNA |
2166 |
0.21 |
| chr7_83653626_83653804 | 0.47 |
Il16 |
interleukin 16 |
1610 |
0.26 |
| chr13_52972166_52972331 | 0.46 |
n-R5s54 |
nuclear encoded rRNA 5S 54 |
7516 |
0.18 |
| chr2_163527264_163527415 | 0.46 |
Gm25881 |
predicted gene, 25881 |
12823 |
0.11 |
| chr4_105459110_105459278 | 0.46 |
Gm12723 |
predicted gene 12723 |
52038 |
0.16 |
| chr17_12392360_12392620 | 0.45 |
Plg |
plasminogen |
13831 |
0.16 |
| chr6_119487150_119487370 | 0.45 |
Fbxl14 |
F-box and leucine-rich repeat protein 14 |
7592 |
0.21 |
| chr18_46669373_46669564 | 0.44 |
Gm3734 |
predicted gene 3734 |
38661 |
0.1 |
| chr16_94693543_94693992 | 0.44 |
Gm41504 |
predicted gene, 41504 |
20136 |
0.16 |
| chr13_93636362_93636873 | 0.44 |
Bhmt |
betaine-homocysteine methyltransferase |
949 |
0.5 |
| chr7_84195894_84196083 | 0.44 |
Gm44826 |
predicted gene 44826 |
14526 |
0.14 |
| chr8_122274769_122274938 | 0.43 |
Zfp469 |
zinc finger protein 469 |
5284 |
0.17 |
| chr10_80101643_80101815 | 0.43 |
Sbno2 |
strawberry notch 2 |
542 |
0.59 |
| chr6_99540166_99540323 | 0.43 |
Foxp1 |
forkhead box P1 |
17523 |
0.21 |
| chr8_33867033_33867186 | 0.43 |
Rbpms |
RNA binding protein gene with multiple splicing |
13436 |
0.16 |
| chr8_121568635_121568786 | 0.43 |
Fbxo31 |
F-box protein 31 |
8747 |
0.11 |
| chr5_92361554_92361705 | 0.42 |
Cxcl11 |
chemokine (C-X-C motif) ligand 11 |
1648 |
0.21 |
| chr15_83568563_83568727 | 0.42 |
Tspo |
translocator protein |
4770 |
0.13 |
| chr9_57233208_57233500 | 0.42 |
Trcg1 |
taste receptor cell gene 1 |
3202 |
0.2 |
| chr4_145016736_145017043 | 0.42 |
Vps13d |
vacuolar protein sorting 13D |
32928 |
0.18 |
| chr6_128095809_128095987 | 0.41 |
Gm26338 |
predicted gene, 26338 |
872 |
0.54 |
| chr12_86797888_86798054 | 0.41 |
Gm10095 |
predicted gene 10095 |
48496 |
0.11 |
| chr9_115342664_115342815 | 0.41 |
Stt3b |
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) |
32318 |
0.11 |
| chr19_20450928_20451527 | 0.41 |
C730002L08Rik |
RIKEN cDNA C730002L08 gene |
26445 |
0.16 |
| chr1_21068954_21069111 | 0.41 |
Tram2 |
translocating chain-associating membrane protein 2 |
10197 |
0.17 |
| chr4_132234596_132234776 | 0.41 |
Gmeb1 |
glucocorticoid modulatory element binding protein 1 |
220 |
0.83 |
| chr19_20467728_20467894 | 0.41 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
24904 |
0.18 |
| chr16_24084566_24084855 | 0.40 |
Gm46545 |
predicted gene, 46545 |
2020 |
0.29 |
| chr8_33799864_33800058 | 0.40 |
Rbpms |
RNA binding protein gene with multiple splicing |
2747 |
0.22 |
| chr8_24448541_24448692 | 0.40 |
Gm44620 |
predicted gene 44620 |
1519 |
0.33 |
| chr1_190121129_190121551 | 0.40 |
Gm28172 |
predicted gene 28172 |
47330 |
0.13 |
| chr5_137610897_137611097 | 0.39 |
Pcolce |
procollagen C-endopeptidase enhancer protein |
295 |
0.7 |
| chr4_141115485_141115659 | 0.39 |
4921514A10Rik |
RIKEN cDNA 4921514A10 gene |
88 |
0.95 |
| chr15_36904059_36904218 | 0.39 |
Gm10384 |
predicted gene 10384 |
24322 |
0.13 |
| chr14_51905961_51906128 | 0.39 |
Ndrg2 |
N-myc downstream regulated gene 2 |
2897 |
0.12 |
| chr12_69366525_69366676 | 0.39 |
Gm18113 |
predicted gene, 18113 |
3423 |
0.12 |
| chr16_76299623_76299791 | 0.38 |
Nrip1 |
nuclear receptor interacting protein 1 |
23951 |
0.22 |
| chr14_58032344_58032511 | 0.38 |
Gm9012 |
predicted gene 9012 |
17396 |
0.15 |
| chr4_144948323_144948474 | 0.38 |
Gm38074 |
predicted gene, 38074 |
10450 |
0.19 |
| chr11_98754990_98755167 | 0.38 |
Thra |
thyroid hormone receptor alpha |
1492 |
0.24 |
| chr1_167354489_167354678 | 0.38 |
Aldh9a1 |
aldehyde dehydrogenase 9, subfamily A1 |
4271 |
0.14 |
| chr17_47444308_47444531 | 0.38 |
1700001C19Rik |
RIKEN cDNA 1700001C19 gene |
7043 |
0.12 |
| chr17_69397260_69397453 | 0.38 |
Gm49894 |
predicted gene, 49894 |
13148 |
0.14 |
| chr5_91740258_91740421 | 0.37 |
Trmt112-ps1 |
tRNA methyltransferase 11-2, pseudogene 1 |
2260 |
0.22 |
| chr4_155604279_155604430 | 0.37 |
Slc35e2 |
solute carrier family 35, member E2 |
2410 |
0.14 |
| chr19_46134425_46134604 | 0.37 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
2617 |
0.16 |
| chr17_26638548_26638713 | 0.37 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
2607 |
0.19 |
| chr3_84262685_84262871 | 0.37 |
Trim2 |
tripartite motif-containing 2 |
2966 |
0.34 |
| chr13_35066144_35066299 | 0.37 |
Gm48629 |
predicted gene, 48629 |
760 |
0.57 |
| chr6_54883509_54883670 | 0.37 |
Gm44185 |
predicted gene, 44185 |
18833 |
0.18 |
| chr6_53154644_53154804 | 0.37 |
Gm8143 |
predicted gene 8143 |
17011 |
0.22 |
| chr15_25647828_25647998 | 0.37 |
Gm48996 |
predicted gene, 48996 |
13286 |
0.18 |
| chr19_3358921_3359072 | 0.37 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
7334 |
0.14 |
| chr14_21422894_21423226 | 0.36 |
Gm25864 |
predicted gene, 25864 |
27414 |
0.17 |
| chr2_91196048_91196252 | 0.36 |
Nr1h3 |
nuclear receptor subfamily 1, group H, member 3 |
999 |
0.35 |
| chr19_41381691_41381880 | 0.36 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
3311 |
0.31 |
| chr16_31165066_31165351 | 0.36 |
Gm18237 |
predicted gene, 18237 |
7338 |
0.18 |
| chr3_121200941_121201336 | 0.36 |
Gm5710 |
predicted gene 5710 |
20041 |
0.13 |
| chr8_70170192_70170368 | 0.36 |
Tmem161a |
transmembrane protein 161A |
2076 |
0.16 |
| chr14_20166542_20166877 | 0.36 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
15100 |
0.13 |
| chr7_65317537_65317718 | 0.36 |
Tjp1 |
tight junction protein 1 |
15322 |
0.21 |
| chr1_118967578_118967755 | 0.36 |
Mir6346 |
microRNA 6346 |
13446 |
0.21 |
| chr3_86018925_86019076 | 0.36 |
Prss48 |
protease, serine 48 |
16509 |
0.12 |
| chr18_20984644_20984795 | 0.35 |
Rnf138 |
ring finger protein 138 |
16622 |
0.19 |
| chr4_96583762_96583984 | 0.35 |
Cyp2j9 |
cytochrome P450, family 2, subfamily j, polypeptide 9 |
7705 |
0.22 |
| chr2_103594624_103594957 | 0.35 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
28480 |
0.17 |
| chr3_27474078_27474242 | 0.35 |
Gm43344 |
predicted gene 43344 |
72481 |
0.1 |
| chr7_127812426_127812775 | 0.35 |
Gm45204 |
predicted gene 45204 |
907 |
0.28 |
| chr10_114798917_114799087 | 0.35 |
Trhde |
TRH-degrading enzyme |
2368 |
0.27 |
| chr11_116126235_116126386 | 0.35 |
Trim47 |
tripartite motif-containing 47 |
900 |
0.38 |
| chr13_59668840_59669190 | 0.35 |
Golm1 |
golgi membrane protein 1 |
6757 |
0.1 |
| chr3_83996881_83997402 | 0.35 |
Tmem131l |
transmembrane 131 like |
28913 |
0.2 |
| chr8_109006416_109006567 | 0.35 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
68731 |
0.11 |
| chr10_120977639_120977804 | 0.35 |
Lemd3 |
LEM domain containing 3 |
1611 |
0.28 |
| chr4_48907830_48908179 | 0.35 |
Gm12436 |
predicted gene 12436 |
9411 |
0.2 |
| chr18_68235108_68235287 | 0.35 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
7130 |
0.19 |
| chr9_108087094_108087536 | 0.34 |
Apeh |
acylpeptide hydrolase |
847 |
0.31 |
| chr1_119818436_119818638 | 0.34 |
Gm8338 |
predicted gene 8338 |
774 |
0.53 |
| chr18_70573730_70573894 | 0.34 |
Mbd2 |
methyl-CpG binding domain protein 2 |
5478 |
0.21 |
| chr5_102643080_102643640 | 0.34 |
Arhgap24 |
Rho GTPase activating protein 24 |
81613 |
0.11 |
| chr3_88833523_88833680 | 0.34 |
1500004A13Rik |
RIKEN cDNA 1500004A13 gene |
1054 |
0.32 |
| chr7_96960854_96961014 | 0.34 |
C230038L03Rik |
RIKEN cDNA C230038L03 gene |
9048 |
0.18 |
| chr4_41630291_41630458 | 0.34 |
Dnaic1 |
dynein, axonemal, intermediate chain 1 |
1270 |
0.29 |
| chr8_120544876_120545045 | 0.34 |
Mir7687 |
microRNA 7687 |
6264 |
0.11 |
| chr19_7196833_7197251 | 0.33 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
3422 |
0.16 |
| chr18_75800139_75800301 | 0.33 |
1700034B16Rik |
RIKEN cDNA 1700034B16 gene |
19188 |
0.18 |
| chr9_65243868_65244187 | 0.33 |
Cilp |
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
21153 |
0.1 |
| chr17_80483459_80483673 | 0.33 |
Sos1 |
SOS Ras/Rac guanine nucleotide exchange factor 1 |
3113 |
0.28 |
| chr1_133012626_133012917 | 0.33 |
Mdm4 |
transformed mouse 3T3 cell double minute 4 |
6087 |
0.16 |
| chr8_120492128_120492303 | 0.33 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
3768 |
0.18 |
| chr15_36903404_36903565 | 0.33 |
Gm10384 |
predicted gene 10384 |
23668 |
0.13 |
| chr2_165896508_165896668 | 0.33 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
362 |
0.81 |
| chr19_44128120_44128295 | 0.32 |
Cwf19l1 |
CWF19-like 1, cell cycle control (S. pombe) |
5291 |
0.13 |
| chr9_22155816_22155994 | 0.32 |
Pigyl |
phosphatidylinositol glycan anchor biosynthesis, class Y-like |
941 |
0.31 |
| chr16_26362547_26362834 | 0.32 |
Cldn1 |
claudin 1 |
1896 |
0.48 |
| chr15_79741187_79741771 | 0.32 |
Sun2 |
Sad1 and UNC84 domain containing 2 |
613 |
0.45 |
| chr19_23002157_23002332 | 0.32 |
Gm50136 |
predicted gene, 50136 |
59210 |
0.12 |
| chr15_78531814_78531981 | 0.32 |
C1qtnf6 |
C1q and tumor necrosis factor related protein 6 |
481 |
0.53 |
| chr17_28010242_28010561 | 0.32 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
3056 |
0.16 |
| chr19_3328497_3328665 | 0.32 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
5144 |
0.14 |
| chr17_44105478_44105637 | 0.32 |
Enpp4 |
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
207 |
0.95 |
| chr12_71041658_71041810 | 0.32 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
6607 |
0.17 |
| chr11_23447377_23447534 | 0.32 |
Usp34 |
ubiquitin specific peptidase 34 |
10483 |
0.15 |
| chr14_41125955_41126187 | 0.32 |
Sftpa1 |
surfactant associated protein A1 |
5711 |
0.13 |
| chr17_11752805_11752972 | 0.31 |
Gm10513 |
predicted gene 10513 |
20543 |
0.26 |
| chr14_47186674_47186843 | 0.31 |
Gch1 |
GTP cyclohydrolase 1 |
2655 |
0.15 |
| chr6_124561569_124561840 | 0.31 |
Gm18840 |
predicted gene, 18840 |
489 |
0.66 |
| chr6_119409619_119409792 | 0.31 |
Adipor2 |
adiponectin receptor 2 |
7770 |
0.21 |
| chr10_87517479_87517681 | 0.31 |
Pah |
phenylalanine hydroxylase |
4215 |
0.22 |
| chr5_102537866_102538044 | 0.30 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
54666 |
0.14 |
| chr1_73992943_73993094 | 0.30 |
Tns1 |
tensin 1 |
2354 |
0.35 |
| chr4_19523139_19523419 | 0.30 |
Cpne3 |
copine III |
20399 |
0.22 |
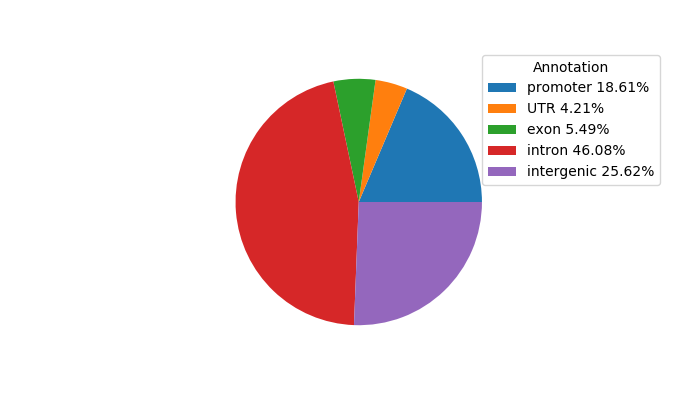
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.8 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.1 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.2 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.4 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.1 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.6 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.3 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.0 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.0 | GO:0072174 | metanephric tubule formation(GO:0072174) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.0 | GO:0009757 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.0 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.0 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.5 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.2 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.2 | 0.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0034785 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0001030 | RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.0 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |