Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
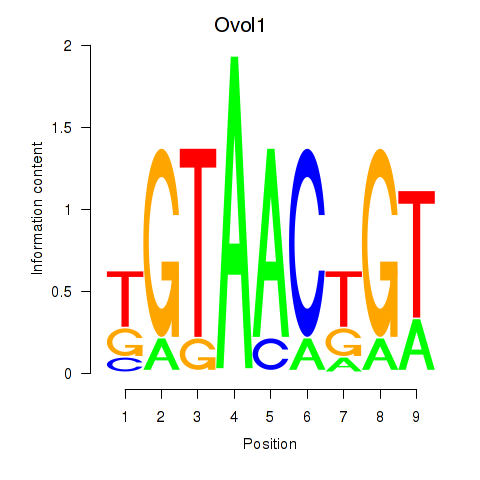
Results for Ovol1
Z-value: 1.98
Transcription factors associated with Ovol1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ovol1
|
ENSMUSG00000024922.1 | ovo like zinc finger 1 |
Activity of the Ovol1 motif across conditions
Conditions sorted by the z-value of the Ovol1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
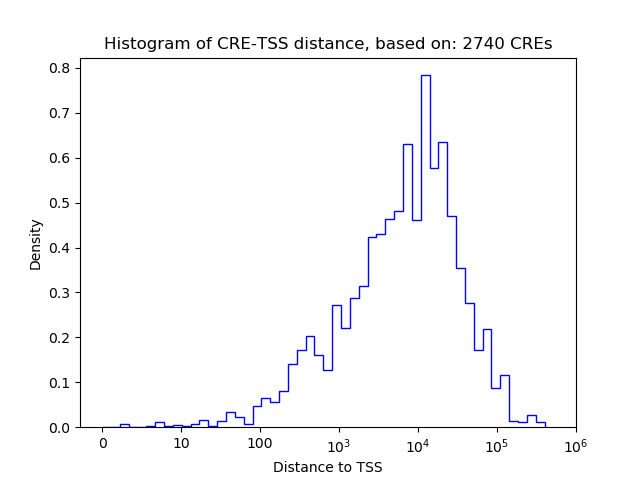
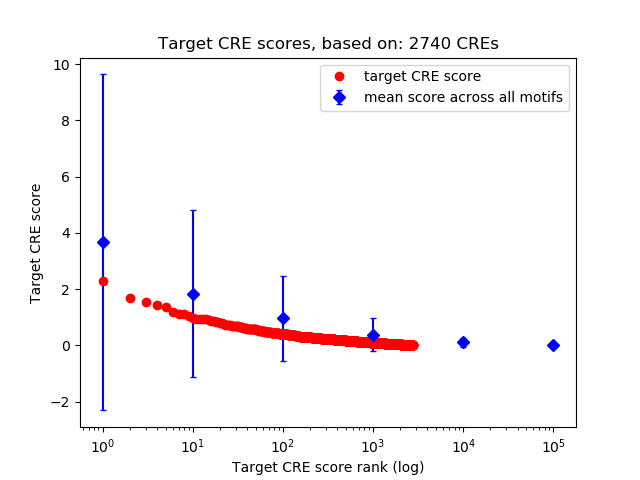
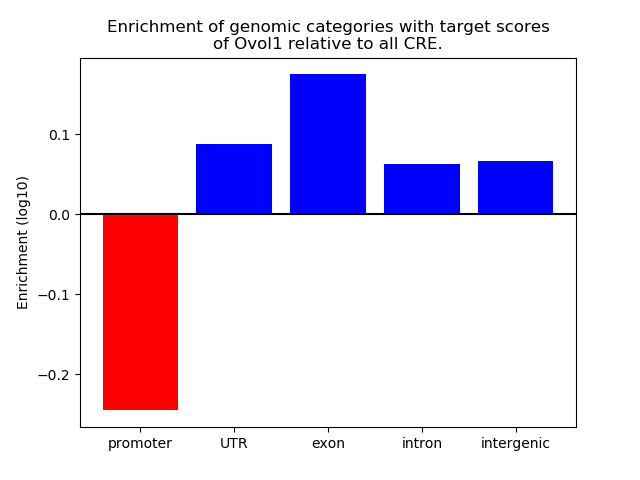
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_186325767_186325937 | 2.30 |
Gm37491 |
predicted gene, 37491 |
21463 |
0.24 |
| chr14_114864274_114864425 | 1.68 |
Gm49010 |
predicted gene, 49010 |
12161 |
0.18 |
| chr16_43348317_43348468 | 1.54 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
14656 |
0.16 |
| chr8_105095287_105095438 | 1.43 |
Ces3b |
carboxylesterase 3B |
6743 |
0.11 |
| chr9_74331100_74331251 | 1.35 |
Gm24141 |
predicted gene, 24141 |
31435 |
0.18 |
| chr9_85551927_85552078 | 1.18 |
Gm48834 |
predicted gene, 48834 |
1763 |
0.42 |
| chr10_99656891_99657084 | 1.13 |
Gm35101 |
predicted gene, 35101 |
12527 |
0.16 |
| chr10_113290132_113290294 | 1.10 |
Gm47532 |
predicted gene, 47532 |
133971 |
0.05 |
| chr12_8081681_8081832 | 1.04 |
Apob |
apolipoprotein B |
69397 |
0.1 |
| chr11_51753632_51753825 | 0.99 |
Sec24a |
Sec24 related gene family, member A (S. cerevisiae) |
2483 |
0.22 |
| chr1_64920991_64921142 | 0.95 |
Plekhm3 |
pleckstrin homology domain containing, family M, member 3 |
7031 |
0.17 |
| chr14_51255648_51255799 | 0.93 |
Rnase2a |
ribonuclease, RNase A family, 2A (liver, eosinophil-derived neurotoxin) |
389 |
0.76 |
| chr1_21245441_21245592 | 0.93 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4887 |
0.13 |
| chr9_74881714_74881880 | 0.92 |
Onecut1 |
one cut domain, family member 1 |
15313 |
0.15 |
| chr16_43287089_43287284 | 0.90 |
Gm37946 |
predicted gene, 37946 |
21095 |
0.17 |
| chr14_62909739_62909890 | 0.87 |
n-R5s47 |
nuclear encoded rRNA 5S 47 |
40100 |
0.09 |
| chr12_32707578_32707729 | 0.87 |
Gm18726 |
predicted gene, 18726 |
2508 |
0.33 |
| chr8_93183570_93183871 | 0.84 |
Gm45909 |
predicted gene 45909 |
7638 |
0.14 |
| chr18_65989569_65989860 | 0.83 |
Lman1 |
lectin, mannose-binding, 1 |
3946 |
0.15 |
| chr5_89352355_89352713 | 0.80 |
Gc |
vitamin D binding protein |
83094 |
0.1 |
| chr17_84650132_84650304 | 0.78 |
Dync2li1 |
dynein cytoplasmic 2 light intermediate chain 1 |
943 |
0.47 |
| chr3_62404931_62405082 | 0.77 |
Arhgef26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
14662 |
0.25 |
| chr2_172975275_172975541 | 0.72 |
Spo11 |
SPO11 meiotic protein covalently bound to DSB |
2292 |
0.24 |
| chr9_74892937_74893252 | 0.72 |
Onecut1 |
one cut domain, family member 1 |
26610 |
0.14 |
| chr15_6891349_6891647 | 0.71 |
Osmr |
oncostatin M receptor |
16529 |
0.26 |
| chr12_40574426_40574700 | 0.71 |
Dock4 |
dedicator of cytokinesis 4 |
128227 |
0.05 |
| chr4_109785016_109785167 | 0.70 |
Gm12808 |
predicted gene 12808 |
30274 |
0.18 |
| chr16_45970303_45970469 | 0.69 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
16788 |
0.15 |
| chr7_80174092_80174243 | 0.69 |
Sema4b |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
12674 |
0.1 |
| chr15_7199123_7199469 | 0.68 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
23773 |
0.22 |
| chr5_62037819_62037970 | 0.68 |
Gm42430 |
predicted gene 42430 |
33674 |
0.23 |
| chr1_67227779_67227978 | 0.68 |
Gm15668 |
predicted gene 15668 |
21322 |
0.2 |
| chr11_16818044_16818409 | 0.67 |
Egfros |
epidermal growth factor receptor, opposite strand |
12476 |
0.22 |
| chr2_146974639_146974805 | 0.66 |
Kiz |
kizuna centrosomal protein |
21839 |
0.2 |
| chr10_117435719_117435921 | 0.66 |
Gm9002 |
predicted gene 9002 |
6202 |
0.15 |
| chr5_123020545_123020720 | 0.63 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
5294 |
0.09 |
| chr18_7657709_7657860 | 0.62 |
Mpp7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
30918 |
0.17 |
| chr17_28982482_28982633 | 0.61 |
Stk38 |
serine/threonine kinase 38 |
371 |
0.72 |
| chr3_27360140_27360291 | 0.60 |
Ghsr |
growth hormone secretagogue receptor |
11136 |
0.21 |
| chr13_62909790_62909949 | 0.59 |
Fbp1 |
fructose bisphosphatase 1 |
21587 |
0.13 |
| chr7_93588215_93588366 | 0.59 |
Gm45039 |
predicted gene 45039 |
20026 |
0.19 |
| chr9_57711329_57711506 | 0.59 |
Edc3 |
enhancer of mRNA decapping 3 |
2850 |
0.18 |
| chr2_121428399_121428550 | 0.59 |
Pdia3 |
protein disulfide isomerase associated 3 |
3844 |
0.09 |
| chr11_16836119_16836554 | 0.59 |
Egfros |
epidermal growth factor receptor, opposite strand |
5634 |
0.23 |
| chr14_25577605_25577757 | 0.57 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
29676 |
0.15 |
| chr11_16818941_16819203 | 0.57 |
Egfros |
epidermal growth factor receptor, opposite strand |
11630 |
0.22 |
| chr8_3048117_3048282 | 0.57 |
Gm44634 |
predicted gene 44634 |
8095 |
0.2 |
| chr16_37902046_37902197 | 0.57 |
Gpr156 |
G protein-coupled receptor 156 |
14375 |
0.14 |
| chr3_129535291_129535442 | 0.57 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
2639 |
0.2 |
| chr10_121796142_121796293 | 0.56 |
Srgap1 |
SLIT-ROBO Rho GTPase activating protein 1 |
10887 |
0.19 |
| chr3_60141611_60141822 | 0.54 |
Gm24382 |
predicted gene, 24382 |
13969 |
0.22 |
| chr8_126932350_126932537 | 0.54 |
Gm26397 |
predicted gene, 26397 |
12537 |
0.14 |
| chr16_87713148_87713314 | 0.53 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
14227 |
0.2 |
| chr7_107574446_107574681 | 0.53 |
Olfml1 |
olfactomedin-like 1 |
6797 |
0.16 |
| chr6_54033623_54033822 | 0.52 |
Chn2 |
chimerin 2 |
5832 |
0.22 |
| chr14_34328663_34328894 | 0.51 |
Glud1 |
glutamate dehydrogenase 1 |
116 |
0.93 |
| chr10_40512569_40512723 | 0.50 |
Gm18671 |
predicted gene, 18671 |
36246 |
0.14 |
| chr2_154868249_154868417 | 0.50 |
Eif2s2 |
eukaryotic translation initiation factor 2, subunit 2 (beta) |
24449 |
0.17 |
| chr13_44291297_44291488 | 0.50 |
Gm29676 |
predicted gene, 29676 |
10943 |
0.21 |
| chr15_3107457_3107608 | 0.50 |
Gm49222 |
predicted gene, 49222 |
8077 |
0.21 |
| chr5_90474479_90475042 | 0.50 |
Alb |
albumin |
12060 |
0.14 |
| chr8_80263585_80263752 | 0.50 |
Gm45430 |
predicted gene 45430 |
68469 |
0.12 |
| chr4_3319048_3319257 | 0.49 |
Gm11786 |
predicted gene 11786 |
8023 |
0.21 |
| chr13_52191366_52191526 | 0.48 |
Gm48199 |
predicted gene, 48199 |
11035 |
0.27 |
| chr15_99038783_99039270 | 0.48 |
Tuba1c |
tubulin, alpha 1C |
8705 |
0.09 |
| chr4_135649660_135649811 | 0.48 |
1700029M20Rik |
RIKEN cDNA 1700029M20 gene |
22482 |
0.13 |
| chr12_51274262_51274425 | 0.47 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
23144 |
0.22 |
| chr18_34937071_34937222 | 0.47 |
Hspa9 |
heat shock protein 9 |
2164 |
0.19 |
| chr7_97411671_97412103 | 0.47 |
Thrsp |
thyroid hormone responsive |
5632 |
0.13 |
| chr10_88784083_88784275 | 0.47 |
Utp20 |
UTP20 small subunit processome component |
1513 |
0.36 |
| chr1_67213400_67213609 | 0.46 |
Gm15668 |
predicted gene 15668 |
35696 |
0.17 |
| chr9_106230886_106231037 | 0.46 |
Alas1 |
aminolevulinic acid synthase 1 |
6123 |
0.1 |
| chr3_51254730_51254881 | 0.46 |
Elf2 |
E74-like factor 2 |
5436 |
0.14 |
| chr15_59447953_59448104 | 0.45 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
73741 |
0.09 |
| chr6_121171179_121171645 | 0.44 |
Pex26 |
peroxisomal biogenesis factor 26 |
12255 |
0.13 |
| chr2_81270139_81270299 | 0.44 |
Gm23900 |
predicted gene, 23900 |
273123 |
0.01 |
| chr19_12717960_12718111 | 0.44 |
Gm15962 |
predicted gene 15962 |
3168 |
0.14 |
| chr6_54465003_54465192 | 0.44 |
Wipf3 |
WAS/WASL interacting protein family, member 3 |
6905 |
0.17 |
| chr1_48564270_48564440 | 0.44 |
Gm28830 |
predicted gene 28830 |
36716 |
0.21 |
| chr17_12453269_12453430 | 0.44 |
C030013G03Rik |
RIKEN cDNA C030013G03 gene |
13495 |
0.19 |
| chr3_69656373_69656571 | 0.44 |
Gm37380 |
predicted gene, 37380 |
5294 |
0.19 |
| chr3_95864904_95865315 | 0.43 |
Mrps21 |
mitochondrial ribosomal protein S21 |
1802 |
0.16 |
| chr8_36753814_36753974 | 0.43 |
Dlc1 |
deleted in liver cancer 1 |
20840 |
0.26 |
| chr11_120815765_120815980 | 0.43 |
Fasn |
fatty acid synthase |
417 |
0.69 |
| chr11_22846775_22846926 | 0.43 |
Gm23772 |
predicted gene, 23772 |
18 |
0.96 |
| chr9_19690640_19690851 | 0.43 |
Gm7769 |
predicted gene 7769 |
126 |
0.94 |
| chr4_117997840_117998088 | 0.43 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
22928 |
0.14 |
| chr15_81481395_81481613 | 0.43 |
Gm49406 |
predicted gene, 49406 |
4877 |
0.14 |
| chr5_113127021_113127172 | 0.43 |
F830115B05Rik |
RIKEN cDNA F830115B05 gene |
7632 |
0.11 |
| chr6_52606460_52606970 | 0.43 |
Gm44434 |
predicted gene, 44434 |
3470 |
0.19 |
| chr10_34131764_34131915 | 0.42 |
Calhm6 |
calcium homeostasis modulator family member 6 |
3855 |
0.16 |
| chr6_119359922_119360073 | 0.42 |
Cacna2d4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
27435 |
0.15 |
| chr11_16756467_16757088 | 0.42 |
Egfr |
epidermal growth factor receptor |
4547 |
0.23 |
| chr15_55043149_55043337 | 0.41 |
Taf2 |
TATA-box binding protein associated factor 2 |
2923 |
0.22 |
| chr2_34779244_34779475 | 0.41 |
Hspa5 |
heat shock protein 5 |
4512 |
0.15 |
| chr1_63112731_63113827 | 0.41 |
Ino80dos |
INO80 complex subunit D, opposite strand |
201 |
0.84 |
| chr1_59687623_59687774 | 0.41 |
Nop58 |
NOP58 ribonucleoprotein |
2481 |
0.14 |
| chr5_4076458_4076777 | 0.41 |
Gm43031 |
predicted gene 43031 |
3046 |
0.24 |
| chr5_23562685_23562882 | 0.41 |
Srpk2 |
serine/arginine-rich protein specific kinase 2 |
13413 |
0.19 |
| chr9_21838672_21838867 | 0.41 |
Angptl8 |
angiopoietin-like 8 |
3259 |
0.14 |
| chr13_112118684_112118873 | 0.41 |
Gm31104 |
predicted gene, 31104 |
19338 |
0.19 |
| chr10_31501811_31501962 | 0.41 |
Gm47687 |
predicted gene, 47687 |
49804 |
0.09 |
| chr14_76409148_76409302 | 0.41 |
Tsc22d1 |
TSC22 domain family, member 1 |
5736 |
0.29 |
| chr19_44393983_44394181 | 0.41 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12608 |
0.14 |
| chr15_7162840_7163002 | 0.40 |
Lifr |
LIF receptor alpha |
8568 |
0.27 |
| chr2_20921163_20921333 | 0.40 |
Arhgap21 |
Rho GTPase activating protein 21 |
6554 |
0.21 |
| chr3_46391803_46391994 | 0.40 |
Gm18843 |
predicted gene, 18843 |
21504 |
0.24 |
| chr3_52648763_52649088 | 0.40 |
Gm10293 |
predicted pseudogene 10293 |
36090 |
0.17 |
| chr3_31408324_31408475 | 0.39 |
Gm38025 |
predicted gene, 38025 |
67387 |
0.12 |
| chr16_42998828_42999221 | 0.39 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43393 |
0.16 |
| chr12_54362251_54362402 | 0.39 |
Gm7550 |
predicted gene 7550 |
28992 |
0.13 |
| chr15_58980885_58981061 | 0.39 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
3163 |
0.21 |
| chr12_30372879_30373073 | 0.39 |
Sntg2 |
syntrophin, gamma 2 |
289 |
0.93 |
| chr11_16817302_16817617 | 0.38 |
Egfros |
epidermal growth factor receptor, opposite strand |
13243 |
0.21 |
| chr13_56571965_56572403 | 0.38 |
2010203P06Rik |
RIKEN cDNA 2010203P06 gene |
23353 |
0.14 |
| chr18_12744753_12744936 | 0.38 |
Gm7599 |
predicted gene 7599 |
1585 |
0.28 |
| chr8_77046087_77046238 | 0.38 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
81851 |
0.09 |
| chr14_116350280_116350464 | 0.37 |
Gm38045 |
predicted gene, 38045 |
399843 |
0.01 |
| chr2_79919674_79919842 | 0.37 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
8033 |
0.3 |
| chr2_4941693_4941844 | 0.37 |
Gm13194 |
predicted gene 13194 |
406 |
0.76 |
| chr6_28779093_28779393 | 0.37 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
2695 |
0.29 |
| chr7_128623495_128623646 | 0.37 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
12210 |
0.12 |
| chr10_24396476_24396638 | 0.37 |
Gm15271 |
predicted gene 15271 |
50933 |
0.14 |
| chr10_30825158_30825321 | 0.37 |
Hey2 |
hairy/enhancer-of-split related with YRPW motif 2 |
16776 |
0.15 |
| chr7_67649875_67650106 | 0.37 |
Ttc23 |
tetratricopeptide repeat domain 23 |
840 |
0.51 |
| chr12_79453000_79453184 | 0.36 |
Rad51b |
RAD51 paralog B |
125739 |
0.05 |
| chr6_24609749_24609900 | 0.36 |
Lmod2 |
leiomodin 2 (cardiac) |
12062 |
0.14 |
| chr9_32718921_32719093 | 0.36 |
Gm27240 |
predicted gene 27240 |
7836 |
0.2 |
| chr2_160361611_160362000 | 0.36 |
Mafb |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
5260 |
0.27 |
| chr13_86111453_86111688 | 0.36 |
Cox7c |
cytochrome c oxidase subunit 7C |
64666 |
0.12 |
| chr17_28241245_28241399 | 0.36 |
Ppard |
peroxisome proliferator activator receptor delta |
8515 |
0.11 |
| chr19_46624218_46624386 | 0.36 |
Wbp1l |
WW domain binding protein 1 like |
901 |
0.49 |
| chr15_64195944_64196287 | 0.35 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
12376 |
0.24 |
| chr5_53202803_53202991 | 0.35 |
Sel1l3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
10418 |
0.2 |
| chr3_9521650_9521991 | 0.35 |
Gm38001 |
predicted gene, 38001 |
25652 |
0.2 |
| chr4_121189641_121189792 | 0.35 |
Rlf |
rearranged L-myc fusion sequence |
1182 |
0.42 |
| chr3_144446211_144446394 | 0.35 |
Gm5857 |
predicted gene 5857 |
18630 |
0.18 |
| chr19_31968064_31968231 | 0.34 |
9130016M20Rik |
RIKEN cDNA 9130016M20 gene |
18063 |
0.21 |
| chr19_47646596_47646802 | 0.34 |
Col17a1 |
collagen, type XVII, alpha 1 |
2896 |
0.17 |
| chr15_38570240_38570408 | 0.34 |
Gm29697 |
predicted gene, 29697 |
3672 |
0.15 |
| chr2_84995821_84996178 | 0.34 |
Prg3 |
proteoglycan 3 |
7784 |
0.12 |
| chr1_59371411_59371704 | 0.34 |
Gm29016 |
predicted gene 29016 |
7373 |
0.21 |
| chr6_129216337_129216553 | 0.33 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
17529 |
0.1 |
| chr3_152762447_152762598 | 0.33 |
Pigk |
phosphatidylinositol glycan anchor biosynthesis, class K |
859 |
0.63 |
| chr14_121072919_121073071 | 0.32 |
Farp1 |
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
29084 |
0.23 |
| chr15_81253437_81253614 | 0.32 |
8430426J06Rik |
RIKEN cDNA 8430426J06 gene |
1663 |
0.31 |
| chr14_69626550_69626701 | 0.32 |
Gm37513 |
predicted gene, 37513 |
10579 |
0.11 |
| chr16_18073808_18074007 | 0.32 |
Dgcr6 |
DiGeorge syndrome critical region gene 6 |
4427 |
0.15 |
| chr15_35876385_35876551 | 0.32 |
Vps13b |
vacuolar protein sorting 13B |
4746 |
0.19 |
| chr12_104025874_104026200 | 0.32 |
Gm37496 |
predicted gene, 37496 |
1610 |
0.23 |
| chr14_18232288_18232439 | 0.32 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
6648 |
0.15 |
| chr19_57258075_57258251 | 0.32 |
4930449E18Rik |
RIKEN cDNA 4930449E18 gene |
15797 |
0.18 |
| chr15_7198099_7198411 | 0.31 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
24814 |
0.22 |
| chr9_30936768_30936954 | 0.31 |
Adamts8 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
5701 |
0.21 |
| chr7_120184451_120184607 | 0.31 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
10671 |
0.13 |
| chr2_134931319_134931796 | 0.31 |
Gm14036 |
predicted gene 14036 |
127608 |
0.05 |
| chr11_16782319_16782716 | 0.31 |
Egfr |
epidermal growth factor receptor |
30287 |
0.16 |
| chr18_3276237_3276415 | 0.31 |
Crem |
cAMP responsive element modulator |
4752 |
0.25 |
| chr8_56292003_56292154 | 0.31 |
Hpgd |
hydroxyprostaglandin dehydrogenase 15 (NAD) |
2507 |
0.34 |
| chr2_31487515_31488471 | 0.31 |
Ass1 |
argininosuccinate synthetase 1 |
9779 |
0.18 |
| chr1_193926856_193927030 | 0.31 |
Gm21362 |
predicted gene, 21362 |
59934 |
0.16 |
| chr17_24868993_24869535 | 0.30 |
Igfals |
insulin-like growth factor binding protein, acid labile subunit |
3267 |
0.1 |
| chr19_45843362_45843693 | 0.30 |
Gm50196 |
predicted gene, 50196 |
90 |
0.96 |
| chr12_69695612_69695763 | 0.30 |
Sos2 |
SOS Ras/Rho guanine nucleotide exchange factor 2 |
13835 |
0.14 |
| chr3_81149765_81149938 | 0.30 |
Gm16000 |
predicted gene 16000 |
109414 |
0.06 |
| chr5_96437978_96438159 | 0.30 |
Gm33050 |
predicted gene, 33050 |
19675 |
0.21 |
| chr9_98434324_98434475 | 0.30 |
Rbp1 |
retinol binding protein 1, cellular |
11438 |
0.2 |
| chr3_130694798_130695032 | 0.30 |
Ostc |
oligosaccharyltransferase complex subunit (non-catalytic) |
1707 |
0.27 |
| chr1_74807738_74808135 | 0.30 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
14573 |
0.11 |
| chr8_46477705_46478047 | 0.30 |
Gm45244 |
predicted gene 45244 |
4286 |
0.17 |
| chr1_140267910_140268090 | 0.30 |
Gm37777 |
predicted gene, 37777 |
1513 |
0.46 |
| chr10_91261198_91261441 | 0.30 |
Gm18705 |
predicted gene, 18705 |
1975 |
0.31 |
| chr2_17192790_17192972 | 0.30 |
Gm13322 |
predicted gene 13322 |
7394 |
0.26 |
| chr8_93107061_93107411 | 0.30 |
Ces1c |
carboxylesterase 1C |
17293 |
0.14 |
| chr7_39841504_39841670 | 0.30 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
43845 |
0.11 |
| chr3_127009818_127009969 | 0.30 |
Gm43750 |
predicted gene 43750 |
908 |
0.45 |
| chr16_42962415_42962640 | 0.30 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
6896 |
0.24 |
| chr5_145793363_145793514 | 0.29 |
Cyp3a44 |
cytochrome P450, family 3, subfamily a, polypeptide 44 |
12436 |
0.15 |
| chr12_38115305_38115462 | 0.29 |
Gm35755 |
predicted gene, 35755 |
25481 |
0.21 |
| chr18_79022725_79022904 | 0.29 |
Setbp1 |
SET binding protein 1 |
86577 |
0.1 |
| chr11_75173089_75173918 | 0.29 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr14_19823120_19823271 | 0.29 |
Rtraf |
RNA transcription, translation and transport factor |
140 |
0.95 |
| chr11_16809967_16810165 | 0.29 |
Egfros |
epidermal growth factor receptor, opposite strand |
20636 |
0.2 |
| chr6_121221807_121221966 | 0.29 |
Gm15856 |
predicted gene 15856 |
4267 |
0.14 |
| chr4_106767670_106767831 | 0.29 |
Acot11 |
acyl-CoA thioesterase 11 |
3806 |
0.18 |
| chr15_7135727_7135925 | 0.29 |
Lifr |
LIF receptor alpha |
4716 |
0.31 |
| chr7_98352590_98353259 | 0.29 |
Tsku |
tsukushi, small leucine rich proteoglycan |
7155 |
0.18 |
| chr15_54578033_54578470 | 0.29 |
Mal2 |
mal, T cell differentiation protein 2 |
7059 |
0.27 |
| chr2_105233444_105233738 | 0.29 |
Them7 |
thioesterase superfamily member 7 |
9249 |
0.24 |
| chr19_32809166_32809317 | 0.28 |
Pten |
phosphatase and tensin homolog |
1984 |
0.43 |
| chr8_18621633_18621799 | 0.28 |
Mcph1 |
microcephaly, primary autosomal recessive 1 |
3847 |
0.2 |
| chr17_26885855_26886147 | 0.28 |
Gm17382 |
predicted gene, 17382 |
174 |
0.88 |
| chr8_28267111_28267301 | 0.28 |
Gm8100 |
predicted gene 8100 |
81366 |
0.1 |
| chr13_74023512_74023688 | 0.28 |
Gm6263 |
predicted gene 6263 |
3466 |
0.15 |
| chr14_61695272_61695482 | 0.28 |
Gm37820 |
predicted gene, 37820 |
11867 |
0.11 |
| chr4_101436475_101436672 | 0.28 |
Ak4 |
adenylate kinase 4 |
10650 |
0.16 |
| chr6_53603997_53604187 | 0.28 |
Creb5 |
cAMP responsive element binding protein 5 |
30716 |
0.2 |
| chr5_67299892_67300051 | 0.28 |
Slc30a9 |
solute carrier family 30 (zinc transporter), member 9 |
6984 |
0.18 |
| chr9_44096490_44096809 | 0.28 |
Usp2 |
ubiquitin specific peptidase 2 |
3975 |
0.08 |
| chr9_64251105_64251328 | 0.28 |
Map2k1 |
mitogen-activated protein kinase kinase 1 |
2415 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.5 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.3 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.2 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.1 | 0.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0045713 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0000237 | leptotene(GO:0000237) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0048370 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.1 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.2 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.0 | 0.0 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.0 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.3 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.0 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.0 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:1903376 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.0 | 0.0 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1904751 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0002176 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0043910 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0034892 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0044466 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |