Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
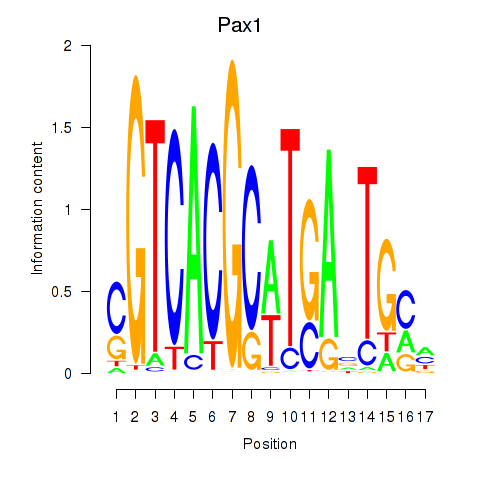
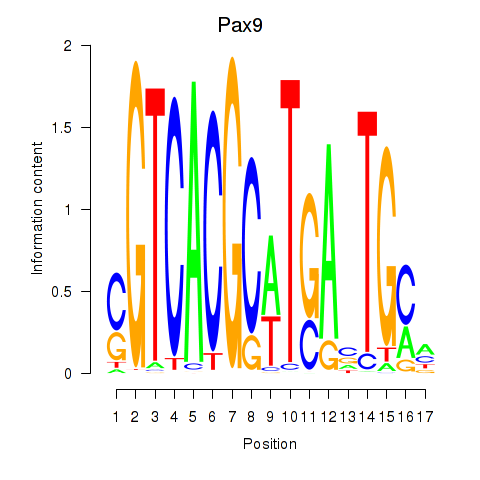
Results for Pax1_Pax9
Z-value: 0.92
Transcription factors associated with Pax1_Pax9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax1
|
ENSMUSG00000037034.9 | paired box 1 |
|
Pax9
|
ENSMUSG00000001497.12 | paired box 9 |
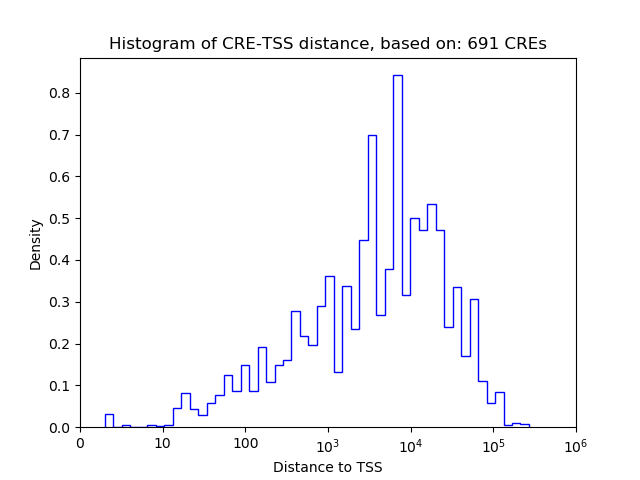
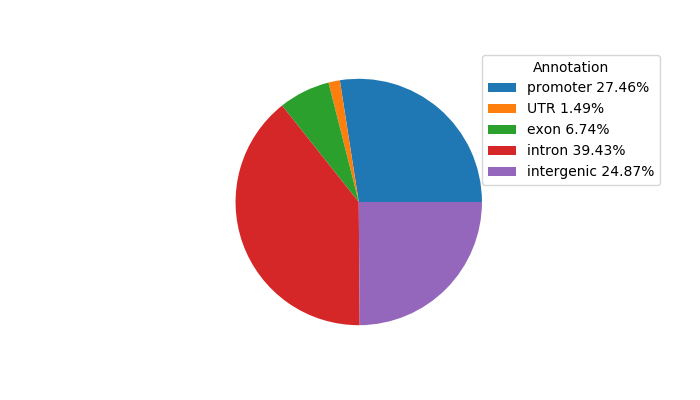
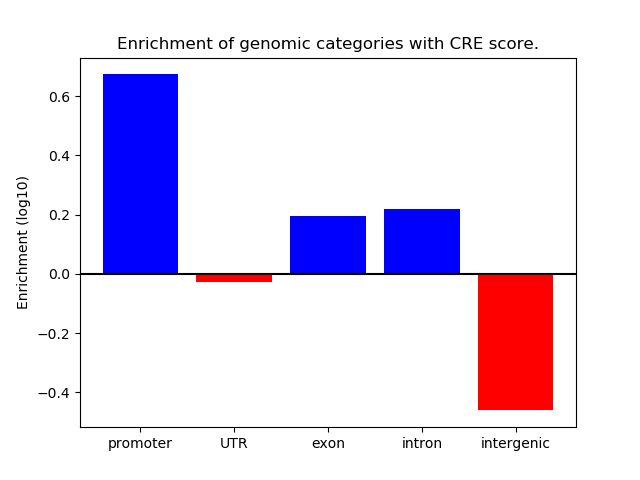
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_147379046_147379216 | Pax1 | 10849 | 0.163052 | -0.76 | 8.2e-02 | Click! |
| chr12_56695474_56695637 | Pax9 | 84 | 0.959601 | 0.72 | 1.0e-01 | Click! |
| chr12_56689847_56690002 | Pax9 | 1843 | 0.264032 | -0.37 | 4.7e-01 | Click! |
| chr12_56681655_56681817 | Pax9 | 10031 | 0.138347 | 0.21 | 6.9e-01 | Click! |
Activity of the Pax1_Pax9 motif across conditions
Conditions sorted by the z-value of the Pax1_Pax9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
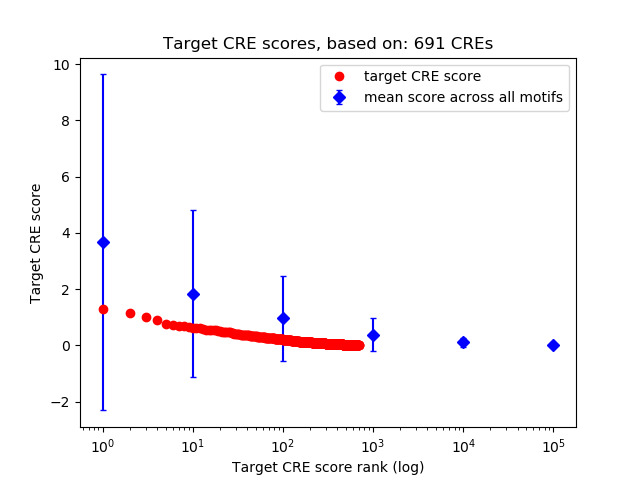
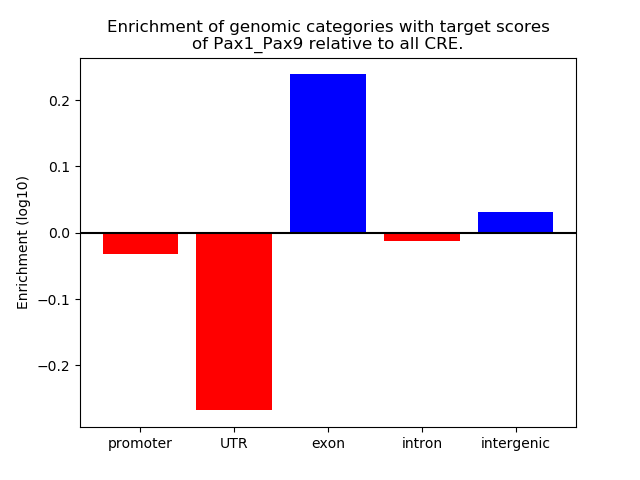
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_138510025_138510190 | 1.30 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
55793 |
0.09 |
| chr3_95652051_95652202 | 1.16 |
Mcl1 |
myeloid cell leukemia sequence 1 |
6662 |
0.1 |
| chr10_61160789_61160964 | 1.01 |
Tbata |
thymus, brain and testes associated |
11078 |
0.15 |
| chr11_94347313_94347820 | 0.89 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
3620 |
0.18 |
| chr2_158225371_158225751 | 0.77 |
D630003M21Rik |
RIKEN cDNA D630003M21 gene |
3661 |
0.17 |
| chr12_83375040_83375279 | 0.73 |
Dpf3 |
D4, zinc and double PHD fingers, family 3 |
24264 |
0.22 |
| chr9_43744780_43745133 | 0.70 |
Nectin1 |
nectin cell adhesion molecule 1 |
312 |
0.63 |
| chr6_128519043_128519655 | 0.67 |
Pzp |
PZP, alpha-2-macroglobulin like |
7354 |
0.09 |
| chr4_150544312_150544715 | 0.64 |
Gm13091 |
predicted gene 13091 |
24031 |
0.18 |
| chr5_120125413_120125579 | 0.62 |
Rbm19 |
RNA binding motif protein 19 |
6900 |
0.2 |
| chr2_25431594_25431764 | 0.61 |
Abca2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
2976 |
0.1 |
| chr2_90547180_90547349 | 0.60 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
33383 |
0.16 |
| chr12_3861728_3861888 | 0.56 |
Dnmt3aos |
DNA methyltransferase 3A, opposite strand |
436 |
0.65 |
| chr6_72637356_72637527 | 0.56 |
Gm37969 |
predicted gene, 37969 |
1114 |
0.24 |
| chr2_84386945_84387393 | 0.55 |
Calcrl |
calcitonin receptor-like |
11811 |
0.2 |
| chr9_120743578_120743729 | 0.55 |
Gm47064 |
predicted gene, 47064 |
7490 |
0.13 |
| chr8_94838256_94838419 | 0.54 |
Coq9 |
coenzyme Q9 |
16 |
0.49 |
| chr9_95573382_95573533 | 0.53 |
Paqr9 |
progestin and adipoQ receptor family member IX |
13800 |
0.12 |
| chr19_24159844_24160184 | 0.52 |
Gm50308 |
predicted gene, 50308 |
5944 |
0.18 |
| chr10_78447521_78447672 | 0.51 |
Pdxk |
pyridoxal (pyridoxine, vitamin B6) kinase |
650 |
0.43 |
| chr5_22345406_22345583 | 0.48 |
Reln |
reelin |
792 |
0.56 |
| chr9_116947417_116947569 | 0.48 |
Gm18489 |
predicted gene, 18489 |
111665 |
0.07 |
| chr5_62622955_62623325 | 0.48 |
Gm43237 |
predicted gene 43237 |
9682 |
0.2 |
| chr2_33468229_33468380 | 0.47 |
Zbtb43 |
zinc finger and BTB domain containing 43 |
201 |
0.93 |
| chr6_92316933_92317084 | 0.46 |
Gm44024 |
predicted gene, 44024 |
35615 |
0.15 |
| chr4_140630998_140631188 | 0.46 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
14028 |
0.17 |
| chr15_78796259_78796421 | 0.46 |
Card10 |
caspase recruitment domain family, member 10 |
5825 |
0.12 |
| chr8_69822995_69823178 | 0.43 |
Lpar2 |
lysophosphatidic acid receptor 2 |
502 |
0.65 |
| chr12_75846819_75846970 | 0.41 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
5836 |
0.27 |
| chr13_54562670_54563000 | 0.41 |
4833439L19Rik |
RIKEN cDNA 4833439L19 gene |
2456 |
0.16 |
| chr18_34328570_34328733 | 0.40 |
Srp19 |
signal recognition particle 19 |
2196 |
0.29 |
| chr17_80833916_80834067 | 0.40 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
62802 |
0.11 |
| chr15_73511980_73512328 | 0.39 |
Dennd3 |
DENN/MADD domain containing 3 |
406 |
0.84 |
| chr11_94328339_94328510 | 0.38 |
Ankrd40 |
ankyrin repeat domain 40 |
170 |
0.93 |
| chr4_60154082_60154314 | 0.38 |
Mup2 |
major urinary protein 2 |
91 |
0.94 |
| chr17_87312523_87312681 | 0.37 |
Ttc7 |
tetratricopeptide repeat domain 7 |
3270 |
0.19 |
| chr4_106577621_106577846 | 0.37 |
Gm12744 |
predicted gene 12744 |
11375 |
0.11 |
| chr16_36711893_36712056 | 0.36 |
Ildr1 |
immunoglobulin-like domain containing receptor 1 |
3564 |
0.17 |
| chr6_141610892_141611057 | 0.36 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
18544 |
0.24 |
| chr11_87442907_87443064 | 0.35 |
Rnu3b1 |
U3B small nuclear RNA 1 |
252 |
0.8 |
| chr6_35303511_35303677 | 0.35 |
Slc13a4 |
solute carrier family 13 (sodium/sulfate symporters), member 4 |
4535 |
0.18 |
| chr5_65421020_65421518 | 0.35 |
Ugdh |
UDP-glucose dehydrogenase |
1537 |
0.22 |
| chr4_135801615_135801933 | 0.35 |
Myom3 |
myomesin family, member 3 |
1130 |
0.41 |
| chr12_3858258_3858439 | 0.35 |
Dnmt3a |
DNA methyltransferase 3A |
2673 |
0.24 |
| chr8_120538771_120538958 | 0.34 |
Mir7687 |
microRNA 7687 |
168 |
0.87 |
| chr1_82281771_82282153 | 0.34 |
Irs1 |
insulin receptor substrate 1 |
9454 |
0.19 |
| chr10_28081351_28081686 | 0.34 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
6508 |
0.24 |
| chr6_90763367_90763528 | 0.34 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
694 |
0.67 |
| chr1_59202432_59202591 | 0.32 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
16615 |
0.14 |
| chr12_17750620_17750832 | 0.32 |
Hpcal1 |
hippocalcin-like 1 |
22500 |
0.2 |
| chr16_4029500_4029663 | 0.32 |
Dnase1 |
deoxyribonuclease I |
7361 |
0.1 |
| chr2_155546997_155547269 | 0.31 |
Gm20637 |
predicted gene 20637 |
748 |
0.41 |
| chr3_131327635_131327824 | 0.31 |
Sgms2 |
sphingomyelin synthase 2 |
17204 |
0.13 |
| chr14_121129398_121129725 | 0.30 |
Farp1 |
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
27482 |
0.23 |
| chr2_163345083_163345234 | 0.30 |
Tox2 |
TOX high mobility group box family member 2 |
24780 |
0.15 |
| chr2_59884229_59884405 | 0.29 |
Wdsub1 |
WD repeat, SAM and U-box domain containing 1 |
1726 |
0.39 |
| chr5_92369721_92369887 | 0.29 |
Cxcl11 |
chemokine (C-X-C motif) ligand 11 |
4319 |
0.12 |
| chr5_66114850_66115151 | 0.29 |
Rbm47 |
RNA binding motif protein 47 |
16809 |
0.11 |
| chr2_33361310_33361488 | 0.29 |
Gm25998 |
predicted gene, 25998 |
1647 |
0.33 |
| chr8_121326131_121326500 | 0.28 |
Gm26815 |
predicted gene, 26815 |
30521 |
0.17 |
| chr15_67062566_67062733 | 0.28 |
Gm31342 |
predicted gene, 31342 |
22591 |
0.2 |
| chr9_46126472_46126634 | 0.28 |
Sik3 |
SIK family kinase 3 |
3316 |
0.24 |
| chr6_119669877_119670185 | 0.28 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
47926 |
0.16 |
| chr3_85022833_85023030 | 0.28 |
Fbxw7 |
F-box and WD-40 domain protein 7 |
70785 |
0.12 |
| chr11_87448290_87448449 | 0.28 |
Rnu3b3 |
U3B small nuclear RNA 3 |
258 |
0.81 |
| chr4_35082276_35082489 | 0.27 |
Ifnk |
interferon kappa |
69674 |
0.09 |
| chr3_118478170_118478340 | 0.27 |
Gm26871 |
predicted gene, 26871 |
20596 |
0.14 |
| chr12_84161882_84162074 | 0.27 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
1840 |
0.21 |
| chr18_64661052_64661203 | 0.26 |
Atp8b1 |
ATPase, class I, type 8B, member 1 |
140 |
0.95 |
| chr8_27197023_27197181 | 0.26 |
Got1l1 |
glutamic-oxaloacetic transaminase 1-like 1 |
1615 |
0.23 |
| chr8_46540344_46540495 | 0.25 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
9355 |
0.15 |
| chr4_61451087_61451318 | 0.25 |
Mup-ps12 |
major urinary protein, pseudogene 12 |
1042 |
0.53 |
| chr4_55014920_55015072 | 0.25 |
Zfp462 |
zinc finger protein 462 |
3516 |
0.32 |
| chr9_61370295_61371248 | 0.25 |
Gm10655 |
predicted gene 10655 |
856 |
0.53 |
| chr9_66511801_66511983 | 0.25 |
Fbxl22 |
F-box and leucine-rich repeat protein 22 |
2717 |
0.23 |
| chr18_53460193_53460360 | 0.25 |
Prdm6 |
PR domain containing 6 |
3307 |
0.33 |
| chr10_96374728_96374886 | 0.25 |
Gm48428 |
predicted gene, 48428 |
28983 |
0.15 |
| chr10_85193452_85193650 | 0.25 |
Cry1 |
cryptochrome 1 (photolyase-like) |
8487 |
0.2 |
| chr17_3121877_3122042 | 0.24 |
Gm49798 |
predicted gene, 49798 |
2842 |
0.14 |
| chr5_32441483_32441679 | 0.24 |
Ppp1cb |
protein phosphatase 1 catalytic subunit beta |
17262 |
0.11 |
| chr13_80893348_80893872 | 0.24 |
Arrdc3 |
arrestin domain containing 3 |
3092 |
0.22 |
| chr2_152396769_152396934 | 0.24 |
Sox12 |
SRY (sex determining region Y)-box 12 |
1195 |
0.26 |
| chr9_20862237_20862476 | 0.24 |
Shfl |
shiftless antiviral inhibitor of ribosomal frameshifting |
6286 |
0.09 |
| chr2_152792514_152792820 | 0.24 |
Gm23802 |
predicted gene, 23802 |
19778 |
0.11 |
| chr9_104140474_104140634 | 0.23 |
Dnajc13 |
DnaJ heat shock protein family (Hsp40) member C13 |
12942 |
0.11 |
| chr13_42829016_42829414 | 0.23 |
Phactr1 |
phosphatase and actin regulator 1 |
37032 |
0.15 |
| chr5_135674436_135674608 | 0.23 |
Por |
P450 (cytochrome) oxidoreductase |
59 |
0.94 |
| chr5_127633759_127633922 | 0.23 |
Slc15a4 |
solute carrier family 15, member 4 |
943 |
0.47 |
| chr1_180161827_180161978 | 0.22 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
895 |
0.53 |
| chr3_41101300_41101479 | 0.22 |
Pgrmc2 |
progesterone receptor membrane component 2 |
18343 |
0.18 |
| chr1_64787740_64787910 | 0.22 |
Plekhm3 |
pleckstrin homology domain containing, family M, member 3 |
48939 |
0.1 |
| chr13_81323663_81323834 | 0.22 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
19088 |
0.24 |
| chr2_90474865_90475228 | 0.22 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
3710 |
0.21 |
| chr18_64633009_64633171 | 0.22 |
Gm6978 |
predicted gene 6978 |
19991 |
0.13 |
| chr15_74921417_74921675 | 0.22 |
Gm6610 |
predicted gene 6610 |
2898 |
0.13 |
| chr5_66000185_66000354 | 0.21 |
9130230L23Rik |
RIKEN cDNA 9130230L23 gene |
51 |
0.96 |
| chr7_44748389_44748555 | 0.21 |
Vrk3 |
vaccinia related kinase 3 |
13 |
0.55 |
| chr4_130120889_130121053 | 0.21 |
Pef1 |
penta-EF hand domain containing 1 |
13415 |
0.14 |
| chr7_37961383_37961554 | 0.21 |
Uri1 |
URI1, prefoldin-like chaperone |
1060 |
0.6 |
| chr1_58718073_58718224 | 0.20 |
Cflar |
CASP8 and FADD-like apoptosis regulator |
4262 |
0.15 |
| chr4_40143379_40143566 | 0.20 |
Aco1 |
aconitase 1 |
391 |
0.86 |
| chr15_10901895_10902215 | 0.20 |
Gm29742 |
predicted gene, 29742 |
15382 |
0.17 |
| chr13_62884266_62884452 | 0.20 |
Fbp1 |
fructose bisphosphatase 1 |
3768 |
0.17 |
| chr3_87982848_87983167 | 0.19 |
Bcan |
brevican |
11448 |
0.09 |
| chr14_30570457_30570647 | 0.19 |
Tkt |
transketolase |
847 |
0.47 |
| chr6_54792866_54793180 | 0.19 |
Znrf2 |
zinc and ring finger 2 |
23893 |
0.19 |
| chr2_32983682_32983870 | 0.19 |
Slc2a8 |
solute carrier family 2, (facilitated glucose transporter), member 8 |
1693 |
0.23 |
| chr2_103811926_103812077 | 0.19 |
Gm13878 |
predicted gene 13878 |
384 |
0.68 |
| chr1_134566100_134566265 | 0.19 |
Kdm5b |
lysine (K)-specific demethylase 5B |
5975 |
0.14 |
| chr4_60755643_60755889 | 0.19 |
Mup-ps8 |
major urinary protein, pseudogene 8 |
1046 |
0.52 |
| chr12_76927037_76927196 | 0.19 |
Max |
Max protein |
12878 |
0.15 |
| chr2_132110806_132111002 | 0.18 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
544 |
0.75 |
| chr19_47067586_47067748 | 0.18 |
Taf5 |
TATA-box binding protein associated factor 5 |
79 |
0.94 |
| chr11_113722506_113722673 | 0.18 |
Cpsf4l |
cleavage and polyadenylation specific factor 4-like |
12572 |
0.13 |
| chr10_13553038_13553200 | 0.18 |
Pex3 |
peroxisomal biogenesis factor 3 |
1 |
0.75 |
| chr13_41341555_41341714 | 0.18 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
17613 |
0.13 |
| chr3_122293242_122293417 | 0.18 |
Gm40190 |
predicted gene, 40190 |
110 |
0.55 |
| chr13_90963501_90963652 | 0.18 |
Rps23 |
ribosomal protein S23 |
39391 |
0.17 |
| chr1_21240367_21240700 | 0.18 |
Gsta3 |
glutathione S-transferase, alpha 3 |
56 |
0.96 |
| chr5_146793134_146793310 | 0.18 |
Usp12 |
ubiquitin specific peptidase 12 |
1784 |
0.25 |
| chr5_138857853_138858235 | 0.17 |
Gm5294 |
predicted gene 5294 |
37964 |
0.15 |
| chr2_160871410_160871577 | 0.17 |
Zhx3 |
zinc fingers and homeoboxes 3 |
1079 |
0.41 |
| chr5_102483708_102483875 | 0.17 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
502 |
0.8 |
| chr18_20749064_20749237 | 0.17 |
B4galt6 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 |
2746 |
0.29 |
| chr9_50845710_50845905 | 0.17 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
506 |
0.74 |
| chr13_66986541_66986692 | 0.17 |
Ptdss1 |
phosphatidylserine synthase 1 |
6852 |
0.12 |
| chr11_50022246_50022426 | 0.17 |
Rnf130 |
ring finger protein 130 |
3010 |
0.24 |
| chr2_51056109_51056315 | 0.17 |
Rnd3 |
Rho family GTPase 3 |
92882 |
0.08 |
| chr9_96258590_96258741 | 0.17 |
Tfdp2 |
transcription factor Dp 2 |
40 |
0.97 |
| chr12_111547368_111547748 | 0.17 |
Eif5 |
eukaryotic translation initiation factor 5 |
4727 |
0.12 |
| chr11_94576436_94576618 | 0.16 |
Acsf2 |
acyl-CoA synthetase family member 2 |
5749 |
0.12 |
| chr16_87524872_87525038 | 0.16 |
Gm24891 |
predicted gene, 24891 |
3210 |
0.17 |
| chrX_168779160_168779328 | 0.16 |
Gm15233 |
predicted gene 15233 |
3988 |
0.27 |
| chr13_96460531_96460839 | 0.16 |
Ankdd1b |
ankyrin repeat and death domain containing 1B |
10426 |
0.14 |
| chr2_119898379_119898542 | 0.16 |
Mga |
MAX gene associated |
1155 |
0.44 |
| chr11_43992113_43992279 | 0.16 |
Gm12153 |
predicted gene 12153 |
33107 |
0.21 |
| chr12_8379850_8380024 | 0.16 |
Gm48075 |
predicted gene, 48075 |
14272 |
0.16 |
| chr1_131092973_131093137 | 0.15 |
Gm37084 |
predicted gene, 37084 |
906 |
0.48 |
| chr2_25224743_25224915 | 0.15 |
Tubb4b |
tubulin, beta 4B class IVB |
127 |
0.85 |
| chr17_64668680_64668845 | 0.15 |
Man2a1 |
mannosidase 2, alpha 1 |
44658 |
0.16 |
| chr12_86793995_86794154 | 0.15 |
Gm10095 |
predicted gene 10095 |
52393 |
0.1 |
| chr8_123727062_123727236 | 0.15 |
6030466F02Rik |
RIKEN cDNA 6030466F02 gene |
6809 |
0.08 |
| chrX_11574471_11574649 | 0.15 |
Gm14515 |
predicted gene 14515 |
26794 |
0.22 |
| chr11_83753876_83754040 | 0.15 |
Heatr6 |
HEAT repeat containing 6 |
71 |
0.94 |
| chr13_69577241_69577408 | 0.15 |
Srd5a1 |
steroid 5 alpha-reductase 1 |
17505 |
0.12 |
| chr2_167888871_167889131 | 0.15 |
Gm14319 |
predicted gene 14319 |
30416 |
0.13 |
| chr5_138979896_138980396 | 0.15 |
Pdgfa |
platelet derived growth factor, alpha |
14135 |
0.17 |
| chr12_21426260_21426445 | 0.14 |
Gm4419 |
predicted gene 4419 |
5593 |
0.17 |
| chr6_90475971_90476128 | 0.14 |
Gm44421 |
predicted gene, 44421 |
2417 |
0.16 |
| chr8_85493343_85493503 | 0.14 |
Gpt2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
814 |
0.6 |
| chr5_138823785_138824155 | 0.14 |
Gm5294 |
predicted gene 5294 |
3890 |
0.23 |
| chr2_70475906_70476144 | 0.13 |
Sp5 |
trans-acting transcription factor 5 |
1102 |
0.41 |
| chr19_53076271_53076445 | 0.13 |
1700054A03Rik |
RIKEN cDNA 1700054A03 gene |
106 |
0.96 |
| chr5_99526510_99527092 | 0.13 |
Gm16227 |
predicted gene 16227 |
16119 |
0.19 |
| chr12_3959908_3960075 | 0.13 |
Pomc |
pro-opiomelanocortin-alpha |
5021 |
0.16 |
| chr15_22046405_22046937 | 0.13 |
Gm49187 |
predicted gene, 49187 |
88239 |
0.1 |
| chr7_79466451_79467019 | 0.13 |
Gm10616 |
predicted gene 10616 |
326 |
0.49 |
| chr11_98935186_98935341 | 0.13 |
Rara |
retinoic acid receptor, alpha |
2435 |
0.17 |
| chr16_23996527_23996692 | 0.13 |
Bcl6 |
B cell leukemia/lymphoma 6 |
7757 |
0.17 |
| chr18_46804511_46804701 | 0.13 |
Gm49972 |
predicted gene, 49972 |
33281 |
0.1 |
| chr6_97278036_97278202 | 0.13 |
Frmd4b |
FERM domain containing 4B |
13142 |
0.18 |
| chr12_26401660_26402061 | 0.13 |
Rnf144a |
ring finger protein 144A |
4587 |
0.18 |
| chr2_172961302_172961478 | 0.13 |
Spo11 |
SPO11 meiotic protein covalently bound to DSB |
16310 |
0.15 |
| chr10_76576688_76577047 | 0.13 |
Ftcd |
formiminotransferase cyclodeaminase |
1215 |
0.33 |
| chr11_109545590_109545763 | 0.13 |
Arsg |
arylsulfatase G |
1922 |
0.3 |
| chr2_77515804_77515970 | 0.13 |
Zfp385b |
zinc finger protein 385B |
3648 |
0.32 |
| chr6_143100150_143100314 | 0.13 |
C2cd5 |
C2 calcium-dependent domain containing 5 |
91 |
0.96 |
| chr15_59046761_59047065 | 0.12 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6316 |
0.23 |
| chr5_122518398_122518573 | 0.12 |
Gm22965 |
predicted gene, 22965 |
11696 |
0.1 |
| chr8_120275994_120276315 | 0.12 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
47698 |
0.12 |
| chr15_64195944_64196287 | 0.12 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
12376 |
0.24 |
| chr1_36536444_36536641 | 0.12 |
Ankrd23 |
ankyrin repeat domain 23 |
21 |
0.94 |
| chr16_24223745_24224269 | 0.12 |
Gm31814 |
predicted gene, 31814 |
7483 |
0.23 |
| chr4_37093076_37093227 | 0.12 |
Taf9-ps |
TATA-box binding protein associated factor 9, pseudogene |
46723 |
0.17 |
| chr5_90464660_90465223 | 0.12 |
Alb |
albumin |
2241 |
0.25 |
| chr5_122802424_122802664 | 0.12 |
Anapc5 |
anaphase-promoting complex subunit 5 |
78 |
0.96 |
| chr7_83892128_83892320 | 0.12 |
Gm49493 |
predicted gene, 49493 |
38 |
0.57 |
| chr9_32540170_32540338 | 0.12 |
Fli1 |
Friend leukemia integration 1 |
1341 |
0.31 |
| chr18_34117842_34118226 | 0.12 |
Gm5238 |
predicted gene 5238 |
7683 |
0.17 |
| chr8_69399199_69399538 | 0.12 |
Gm25661 |
predicted gene, 25661 |
357 |
0.82 |
| chr2_83648966_83649145 | 0.11 |
Zc3h15 |
zinc finger CCCH-type containing 15 |
4432 |
0.24 |
| chr1_69634071_69634222 | 0.11 |
Gm28112 |
predicted gene 28112 |
27915 |
0.17 |
| chr12_112586365_112586574 | 0.11 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
2315 |
0.23 |
| chr10_111795867_111796218 | 0.11 |
Gm47865 |
predicted gene, 47865 |
3709 |
0.16 |
| chr11_72940971_72941129 | 0.11 |
Gm44467 |
predicted gene, 44467 |
12057 |
0.13 |
| chr3_39944706_39944868 | 0.11 |
Gm42785 |
predicted gene 42785 |
121210 |
0.06 |
| chr10_75789893_75790050 | 0.11 |
Gstt1 |
glutathione S-transferase, theta 1 |
7598 |
0.09 |
| chr4_100377331_100377511 | 0.11 |
Gm12706 |
predicted gene 12706 |
15362 |
0.26 |
| chr19_47511131_47511282 | 0.11 |
Gm19557 |
predicted gene, 19557 |
1776 |
0.29 |
| chr13_24049473_24049634 | 0.11 |
Carmil1 |
capping protein regulator and myosin 1 linker 1 |
9766 |
0.17 |
| chr9_77768228_77768406 | 0.11 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
2175 |
0.24 |
| chr14_11591449_11591619 | 0.11 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
37953 |
0.17 |
| chr5_129019617_129019788 | 0.11 |
Ran |
RAN, member RAS oncogene family |
367 |
0.88 |
| chr17_46725800_46726019 | 0.11 |
Gnmt |
glycine N-methyltransferase |
793 |
0.43 |
| chr11_45178287_45178504 | 0.10 |
Gm38087 |
predicted gene, 38087 |
11477 |
0.2 |
| chr5_117380230_117380381 | 0.10 |
Rfc5 |
replication factor C (activator 1) 5 |
89 |
0.95 |
| chr19_41277479_41277665 | 0.10 |
Tm9sf3 |
transmembrane 9 superfamily member 3 |
13575 |
0.24 |
| chr4_60516308_60516518 | 0.10 |
Mup-ps6 |
major urinary protein, pseudogene 6 |
1048 |
0.41 |
| chr19_6340387_6340875 | 0.10 |
Map4k2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
504 |
0.46 |
| chr19_41332343_41332496 | 0.10 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
52677 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.1 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:2000591 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.2 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0034548 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.0 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |