Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
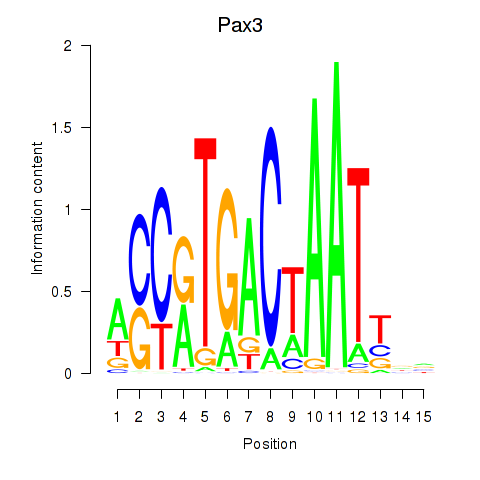
Results for Pax3
Z-value: 1.36
Transcription factors associated with Pax3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax3
|
ENSMUSG00000004872.9 | paired box 3 |
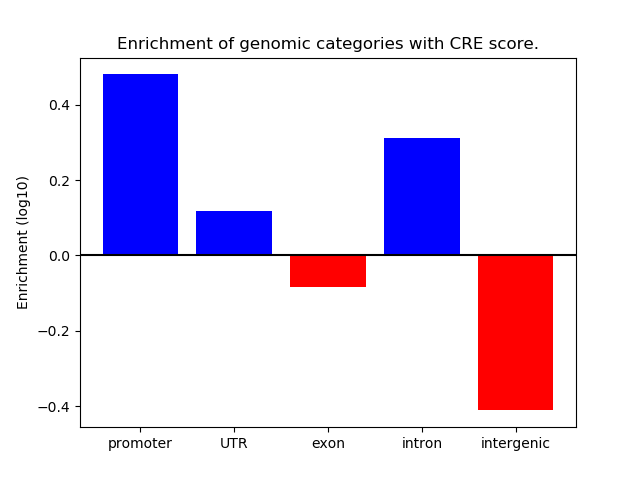
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_78140703_78141066 | Pax3 | 55954 | 0.136991 | 0.81 | 5.2e-02 | Click! |
| chr1_78093772_78093929 | Pax3 | 102988 | 0.067501 | 0.76 | 8.1e-02 | Click! |
| chr1_78142712_78143028 | Pax3 | 53968 | 0.141145 | -0.59 | 2.2e-01 | Click! |
| chr1_78143284_78143452 | Pax3 | 53470 | 0.142201 | 0.59 | 2.2e-01 | Click! |
| chr1_78143066_78143229 | Pax3 | 53691 | 0.141731 | 0.45 | 3.6e-01 | Click! |
Activity of the Pax3 motif across conditions
Conditions sorted by the z-value of the Pax3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
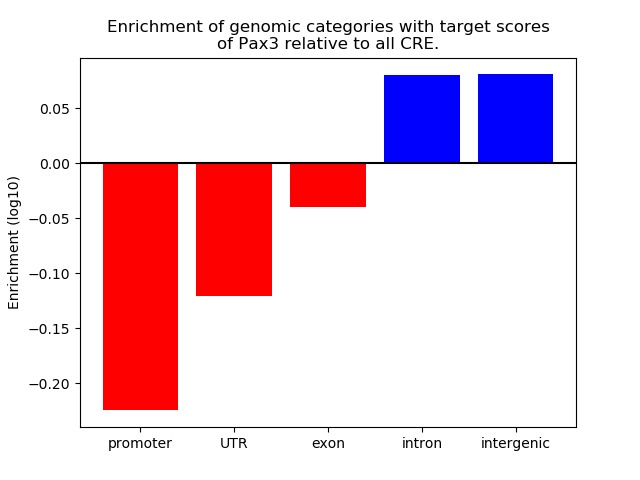
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_30953629_30953810 | 0.86 |
Gm46565 |
predicted gene, 46565 |
18090 |
0.15 |
| chr1_40217416_40217567 | 0.83 |
Il1r1 |
interleukin 1 receptor, type I |
7589 |
0.21 |
| chr7_107574446_107574681 | 0.80 |
Olfml1 |
olfactomedin-like 1 |
6797 |
0.16 |
| chr18_11858447_11858616 | 0.68 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
14231 |
0.16 |
| chr2_134489871_134490022 | 0.64 |
Hao1 |
hydroxyacid oxidase 1, liver |
64361 |
0.14 |
| chr11_109734187_109734610 | 0.62 |
Fam20a |
family with sequence similarity 20, member A |
12119 |
0.17 |
| chr15_7194329_7194480 | 0.53 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
28665 |
0.2 |
| chr12_33285610_33285770 | 0.51 |
Atxn7l1 |
ataxin 7-like 1 |
16825 |
0.2 |
| chr2_115736424_115736575 | 0.51 |
Mir1951 |
microRNA 1951 |
97774 |
0.08 |
| chr10_75051471_75051652 | 0.47 |
Bcr |
BCR activator of RhoGEF and GTPase |
9031 |
0.15 |
| chr2_4895714_4895931 | 0.47 |
Sephs1 |
selenophosphate synthetase 1 |
11438 |
0.15 |
| chr13_56354793_56354953 | 0.46 |
Gm10782 |
predicted gene 10782 |
8027 |
0.15 |
| chr3_127891231_127891382 | 0.43 |
Fam241a |
family with sequence similarity 241, member A |
4982 |
0.14 |
| chr5_125522928_125523080 | 0.42 |
Aacs |
acetoacetyl-CoA synthetase |
7761 |
0.16 |
| chr8_115089778_115090403 | 0.42 |
Gm22556 |
predicted gene, 22556 |
37177 |
0.23 |
| chr6_51702213_51702384 | 0.41 |
Gm38811 |
predicted gene, 38811 |
8783 |
0.25 |
| chr19_37213872_37214035 | 0.41 |
Marchf5 |
membrane associated ring-CH-type finger 5 |
3237 |
0.17 |
| chr16_70320876_70321204 | 0.41 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
6906 |
0.26 |
| chr15_81432149_81432317 | 0.41 |
Gm17025 |
predicted gene 17025 |
13045 |
0.11 |
| chr15_41494169_41494320 | 0.41 |
Oxr1 |
oxidation resistance 1 |
45217 |
0.2 |
| chr1_67234188_67234342 | 0.39 |
Gm15668 |
predicted gene 15668 |
14935 |
0.21 |
| chr19_40156899_40157166 | 0.38 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
30254 |
0.13 |
| chr15_67780972_67781123 | 0.38 |
Gm49408 |
predicted gene, 49408 |
92781 |
0.09 |
| chr5_63953741_63953922 | 0.38 |
Rell1 |
RELT-like 1 |
14992 |
0.15 |
| chr9_71694307_71694467 | 0.38 |
Cgnl1 |
cingulin-like 1 |
46037 |
0.14 |
| chr17_32270584_32270735 | 0.38 |
Brd4 |
bromodomain containing 4 |
1706 |
0.26 |
| chr4_49544305_49544456 | 0.38 |
Aldob |
aldolase B, fructose-bisphosphate |
4494 |
0.16 |
| chr1_93029521_93029757 | 0.37 |
Kif1a |
kinesin family member 1A |
2434 |
0.2 |
| chr13_37537164_37537329 | 0.37 |
Lncbate6 |
brown adipose tissue enriched long noncoding RNA 6 |
4111 |
0.13 |
| chr2_168314984_168315140 | 0.37 |
Gm14262 |
predicted gene 14262 |
19941 |
0.13 |
| chr15_84707077_84707228 | 0.36 |
Arhgap8 |
Rho GTPase activating protein 8 |
12900 |
0.16 |
| chr5_32441483_32441679 | 0.36 |
Ppp1cb |
protein phosphatase 1 catalytic subunit beta |
17262 |
0.11 |
| chr6_51698327_51698491 | 0.36 |
Gm38811 |
predicted gene, 38811 |
12672 |
0.24 |
| chr5_90431005_90431358 | 0.36 |
Alb |
albumin |
29716 |
0.13 |
| chr7_35127036_35127350 | 0.35 |
Gm45091 |
predicted gene 45091 |
7588 |
0.1 |
| chr14_16483661_16483812 | 0.35 |
Top2b |
topoisomerase (DNA) II beta |
58308 |
0.12 |
| chr6_54199490_54199996 | 0.35 |
Gm15526 |
predicted gene 15526 |
22060 |
0.17 |
| chr8_128654178_128654469 | 0.35 |
Gm45746 |
predicted gene 45746 |
10 |
0.97 |
| chr11_100493828_100493979 | 0.35 |
Acly |
ATP citrate lyase |
296 |
0.81 |
| chr2_165825114_165825267 | 0.34 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
26932 |
0.14 |
| chr13_90896944_90897260 | 0.34 |
Atp6ap1l |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
1722 |
0.38 |
| chr17_24116369_24116555 | 0.34 |
Pdpk1 |
3-phosphoinositide dependent protein kinase 1 |
4332 |
0.11 |
| chr15_11505228_11505379 | 0.33 |
Tars |
threonyl-tRNA synthetase |
105638 |
0.06 |
| chr7_71708283_71708442 | 0.32 |
Gm44688 |
predicted gene 44688 |
2286 |
0.26 |
| chr8_4837839_4838020 | 0.32 |
Gm44961 |
predicted gene 44961 |
16699 |
0.13 |
| chr10_105404051_105404220 | 0.32 |
Gm48203 |
predicted gene, 48203 |
9849 |
0.19 |
| chr10_4112716_4112889 | 0.32 |
Gm25515 |
predicted gene, 25515 |
8489 |
0.22 |
| chr11_16830831_16831057 | 0.32 |
Egfros |
epidermal growth factor receptor, opposite strand |
242 |
0.94 |
| chr14_63223276_63223490 | 0.32 |
Gata4 |
GATA binding protein 4 |
9776 |
0.16 |
| chr5_52360112_52360263 | 0.32 |
Sod3 |
superoxide dismutase 3, extracellular |
3604 |
0.19 |
| chr6_52606460_52606970 | 0.31 |
Gm44434 |
predicted gene, 44434 |
3470 |
0.19 |
| chr4_119589534_119589697 | 0.31 |
Foxj3 |
forkhead box J3 |
31727 |
0.1 |
| chr13_56613401_56613633 | 0.31 |
Tgfbi |
transforming growth factor, beta induced |
3931 |
0.22 |
| chr1_62019757_62019954 | 0.31 |
Gm29640 |
predicted gene 29640 |
38648 |
0.18 |
| chr1_39731626_39731803 | 0.31 |
Rfx8 |
regulatory factor X 8 |
10717 |
0.2 |
| chr1_37026954_37027105 | 0.31 |
Vwa3b |
von Willebrand factor A domain containing 3B |
433 |
0.82 |
| chr12_84161882_84162074 | 0.31 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
1840 |
0.21 |
| chr2_58790704_58790855 | 0.30 |
Upp2 |
uridine phosphorylase 2 |
25454 |
0.18 |
| chr3_57696895_57697058 | 0.29 |
Gm43098 |
predicted gene 43098 |
500 |
0.68 |
| chr9_110676644_110676946 | 0.29 |
Gm35715 |
predicted gene, 35715 |
1841 |
0.21 |
| chr4_42805039_42805190 | 0.29 |
Ccl21a |
chemokine (C-C motif) ligand 21A (serine) |
31121 |
0.1 |
| chr11_44461636_44461815 | 0.29 |
Ublcp1 |
ubiquitin-like domain containing CTD phosphatase 1 |
372 |
0.84 |
| chr14_114875214_114875365 | 0.29 |
Gm49010 |
predicted gene, 49010 |
1221 |
0.47 |
| chr2_11598213_11598381 | 0.29 |
Rbm17 |
RNA binding motif protein 17 |
4965 |
0.15 |
| chr19_18181186_18181345 | 0.28 |
Gm18610 |
predicted gene, 18610 |
24530 |
0.25 |
| chr14_33018854_33019018 | 0.28 |
Wdfy4 |
WD repeat and FYVE domain containing 4 |
4401 |
0.24 |
| chr7_35127993_35128152 | 0.28 |
Gm45091 |
predicted gene 45091 |
8467 |
0.1 |
| chr3_108830802_108830953 | 0.28 |
Stxbp3 |
syntaxin binding protein 3 |
9574 |
0.15 |
| chr5_64389400_64389576 | 0.28 |
Gm43837 |
predicted gene 43837 |
6034 |
0.16 |
| chr9_46049413_46049714 | 0.28 |
Sik3 |
SIK family kinase 3 |
36563 |
0.11 |
| chr9_70318576_70318770 | 0.28 |
Mir5626 |
microRNA 5626 |
87080 |
0.07 |
| chr18_7657901_7658087 | 0.28 |
Mpp7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
31128 |
0.17 |
| chr5_110857209_110857384 | 0.28 |
Chek2 |
checkpoint kinase 2 |
8975 |
0.11 |
| chr2_50332059_50332279 | 0.28 |
Gm13483 |
predicted gene 13483 |
35264 |
0.15 |
| chr7_44473035_44473343 | 0.28 |
5430431A17Rik |
RIKEN cDNA 5430431A17 gene |
349 |
0.67 |
| chr7_6244451_6244617 | 0.28 |
Epp13 |
epididymal protein 13 |
8176 |
0.11 |
| chr2_4873838_4873989 | 0.27 |
Sephs1 |
selenophosphate synthetase 1 |
7651 |
0.17 |
| chr1_153741335_153741540 | 0.27 |
Rgs16 |
regulator of G-protein signaling 16 |
1088 |
0.32 |
| chr14_69628830_69628981 | 0.27 |
Gm27177 |
predicted gene 27177 |
11369 |
0.11 |
| chr11_3191105_3191256 | 0.27 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
2124 |
0.19 |
| chr2_119408322_119408473 | 0.27 |
Ino80 |
INO80 complex subunit |
31283 |
0.12 |
| chr11_23459880_23460062 | 0.27 |
Usp34 |
ubiquitin specific peptidase 34 |
1894 |
0.26 |
| chr16_70352880_70353045 | 0.27 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
38828 |
0.19 |
| chr7_104400820_104400986 | 0.27 |
Trim30c |
tripartite motif-containing 30C |
66 |
0.94 |
| chr14_69410565_69410723 | 0.26 |
Gm38378 |
predicted gene, 38378 |
12873 |
0.11 |
| chr12_84762512_84762714 | 0.26 |
Npc2 |
NPC intracellular cholesterol transporter 2 |
73 |
0.96 |
| chr10_64071704_64071882 | 0.26 |
Lrrtm3 |
leucine rich repeat transmembrane neuronal 3 |
18454 |
0.28 |
| chr15_83202527_83202771 | 0.26 |
7530414M10Rik |
RIKEN cDNA 7530414M10 gene |
26204 |
0.1 |
| chr7_81641263_81641414 | 0.26 |
Homer2 |
homer scaffolding protein 2 |
22433 |
0.11 |
| chr5_125487930_125488123 | 0.26 |
Gm27551 |
predicted gene, 27551 |
8649 |
0.13 |
| chr10_111364307_111364473 | 0.25 |
Gm40761 |
predicted gene, 40761 |
27266 |
0.17 |
| chr16_34961005_34961156 | 0.25 |
E130310I04Rik |
RIKEN cDNA E130310I04 gene |
2086 |
0.28 |
| chr2_15112887_15113050 | 0.25 |
Gm13313 |
predicted gene 13313 |
14632 |
0.17 |
| chr12_99426306_99426461 | 0.25 |
Foxn3 |
forkhead box N3 |
13778 |
0.17 |
| chr13_109599464_109599649 | 0.25 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
18151 |
0.3 |
| chr9_30613779_30614132 | 0.25 |
Gm47716 |
predicted gene, 47716 |
18632 |
0.24 |
| chr19_23170256_23170407 | 0.24 |
Mir1192 |
microRNA 1192 |
20900 |
0.15 |
| chr6_50305009_50305231 | 0.24 |
Osbpl3 |
oxysterol binding protein-like 3 |
4766 |
0.27 |
| chr19_10895844_10896002 | 0.24 |
Prpf19 |
pre-mRNA processing factor 19 |
509 |
0.64 |
| chr6_7876922_7877112 | 0.24 |
Gm22276 |
predicted gene, 22276 |
2546 |
0.22 |
| chr10_20739880_20740054 | 0.23 |
Pde7b |
phosphodiesterase 7B |
14889 |
0.2 |
| chr16_95579480_95579639 | 0.23 |
Erg |
ETS transcription factor |
7034 |
0.29 |
| chr6_93348262_93348414 | 0.23 |
Gm25094 |
predicted gene, 25094 |
12750 |
0.26 |
| chr3_131105917_131106088 | 0.23 |
Lef1 |
lymphoid enhancer binding factor 1 |
4469 |
0.19 |
| chr1_106309746_106309897 | 0.23 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
7417 |
0.25 |
| chr19_40164076_40164449 | 0.23 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
23024 |
0.14 |
| chr1_90963133_90963297 | 0.23 |
Rab17 |
RAB17, member RAS oncogene family |
4402 |
0.16 |
| chr9_65196176_65196352 | 0.23 |
Gm25313 |
predicted gene, 25313 |
423 |
0.64 |
| chr5_90471470_90471733 | 0.23 |
Alb |
albumin |
8901 |
0.15 |
| chr10_87858971_87859139 | 0.22 |
Igf1 |
insulin-like growth factor 1 |
13 |
0.98 |
| chr8_115935040_115935191 | 0.22 |
Gm45733 |
predicted gene 45733 |
36956 |
0.23 |
| chr16_32645465_32645635 | 0.22 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
591 |
0.68 |
| chr5_145465006_145465183 | 0.22 |
Cyp3a16 |
cytochrome P450, family 3, subfamily a, polypeptide 16 |
4629 |
0.2 |
| chr19_38372543_38372709 | 0.22 |
Gm50155 |
predicted gene, 50155 |
12888 |
0.14 |
| chr3_82110699_82110861 | 0.22 |
Mir7010 |
microRNA 7010 |
4471 |
0.22 |
| chr1_164632885_164633038 | 0.22 |
Gm37754 |
predicted gene, 37754 |
1204 |
0.46 |
| chr5_91214401_91214579 | 0.22 |
Gm23092 |
predicted gene, 23092 |
3514 |
0.28 |
| chr4_87880355_87880506 | 0.22 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
5888 |
0.31 |
| chr2_25224743_25224915 | 0.22 |
Tubb4b |
tubulin, beta 4B class IVB |
127 |
0.85 |
| chr15_91203650_91203811 | 0.22 |
CN725425 |
cDNA sequence CN725425 |
4236 |
0.24 |
| chr1_64802706_64802963 | 0.22 |
Plekhm3 |
pleckstrin homology domain containing, family M, member 3 |
33930 |
0.14 |
| chr5_33467286_33467498 | 0.22 |
Gm43851 |
predicted gene 43851 |
29928 |
0.14 |
| chr6_72694339_72694529 | 0.22 |
Gm15402 |
predicted gene 15402 |
4780 |
0.14 |
| chr13_85126519_85126671 | 0.21 |
Gm4076 |
predicted gene 4076 |
919 |
0.57 |
| chr9_71217044_71217463 | 0.21 |
Aldh1a2 |
aldehyde dehydrogenase family 1, subfamily A2 |
1464 |
0.43 |
| chr8_121943229_121943385 | 0.21 |
Car5a |
carbonic anhydrase 5a, mitochondrial |
1597 |
0.22 |
| chr9_22800849_22801004 | 0.21 |
Gm27639 |
predicted gene, 27639 |
28091 |
0.22 |
| chr4_8361870_8362071 | 0.21 |
Gm37386 |
predicted gene, 37386 |
92277 |
0.08 |
| chr11_70713419_70713657 | 0.21 |
Mir6925 |
microRNA 6925 |
7548 |
0.08 |
| chr8_48517142_48517293 | 0.21 |
Tenm3 |
teneurin transmembrane protein 3 |
38096 |
0.21 |
| chr9_25526348_25526499 | 0.21 |
Gm25861 |
predicted gene, 25861 |
12782 |
0.19 |
| chr6_31339858_31340224 | 0.21 |
Gm14532 |
predicted gene 14532 |
16702 |
0.14 |
| chr5_115775514_115775665 | 0.21 |
Gm13841 |
predicted gene 13841 |
26570 |
0.14 |
| chr18_70440225_70440381 | 0.21 |
Gm45879 |
predicted gene 45879 |
6447 |
0.19 |
| chr10_125490636_125490927 | 0.21 |
Gm9102 |
predicted gene 9102 |
21750 |
0.25 |
| chr10_43896146_43896340 | 0.21 |
Qrsl1 |
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
702 |
0.66 |
| chr13_41813363_41813628 | 0.21 |
1700061E18Rik |
RIKEN cDNA 1700061E18 gene |
12712 |
0.15 |
| chr3_101601625_101602386 | 0.21 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
2679 |
0.24 |
| chr6_128549526_128549695 | 0.21 |
A2ml1 |
alpha-2-macroglobulin like 1 |
3668 |
0.12 |
| chr13_59498313_59498500 | 0.21 |
Agtpbp1 |
ATP/GTP binding protein 1 |
15748 |
0.16 |
| chr7_14413903_14414054 | 0.20 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
22469 |
0.14 |
| chr5_148777254_148777431 | 0.20 |
4930505K14Rik |
RIKEN cDNA 4930505K14 gene |
621 |
0.67 |
| chr8_91593939_91594090 | 0.20 |
Gm45294 |
predicted gene 45294 |
14726 |
0.14 |
| chr8_78021619_78021802 | 0.20 |
Gm29895 |
predicted gene, 29895 |
22300 |
0.24 |
| chr11_108309880_108310049 | 0.20 |
Apoh |
apolipoprotein H |
33390 |
0.16 |
| chr15_10985300_10985768 | 0.20 |
Amacr |
alpha-methylacyl-CoA racemase |
2128 |
0.26 |
| chr14_30011884_30012221 | 0.20 |
Chdh |
choline dehydrogenase |
2881 |
0.15 |
| chr2_31500949_31501119 | 0.20 |
Ass1 |
argininosuccinate synthetase 1 |
308 |
0.89 |
| chr14_120705561_120705745 | 0.20 |
Rpl19-ps5 |
ribosomal protein L19, pseudogene 5 |
7425 |
0.21 |
| chr10_95837027_95837208 | 0.20 |
Gm33543 |
predicted gene, 33543 |
5779 |
0.13 |
| chr19_20609347_20609744 | 0.20 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
7584 |
0.22 |
| chr4_107999051_107999218 | 0.20 |
Slc1a7 |
solute carrier family 1 (glutamate transporter), member 7 |
10883 |
0.13 |
| chr12_40034619_40034798 | 0.20 |
Arl4a |
ADP-ribosylation factor-like 4A |
2659 |
0.25 |
| chr2_74015081_74015248 | 0.20 |
Gm13668 |
predicted gene 13668 |
54495 |
0.12 |
| chr17_29496086_29496540 | 0.20 |
Pim1 |
proviral integration site 1 |
2906 |
0.15 |
| chr2_145822849_145823024 | 0.20 |
Rin2 |
Ras and Rab interactor 2 |
647 |
0.75 |
| chr3_86013918_86014185 | 0.19 |
Prss48 |
protease, serine 48 |
11560 |
0.13 |
| chr10_127970687_127970872 | 0.19 |
Gm47949 |
predicted gene, 47949 |
9223 |
0.09 |
| chr2_91631313_91631664 | 0.19 |
F2 |
coagulation factor II |
4907 |
0.12 |
| chr5_116435060_116435476 | 0.19 |
Srrm4os |
serine/arginine repetitive matrix 4, opposite strand |
3452 |
0.16 |
| chr15_79685106_79685264 | 0.19 |
Josd1 |
Josephin domain containing 1 |
2734 |
0.12 |
| chr2_120540080_120540275 | 0.19 |
Zfp106 |
zinc finger protein 106 |
317 |
0.86 |
| chr2_122258184_122258335 | 0.19 |
Sord |
sorbitol dehydrogenase |
23510 |
0.09 |
| chr12_3846090_3846255 | 0.19 |
Dnmt3a |
DNA methyltransferase 3A |
3463 |
0.22 |
| chr11_38679459_38679621 | 0.19 |
Gm23520 |
predicted gene, 23520 |
118737 |
0.07 |
| chr1_157507362_157507522 | 0.19 |
Sec16b |
SEC16 homolog B (S. cerevisiae) |
632 |
0.67 |
| chr10_28524122_28524273 | 0.19 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
35954 |
0.22 |
| chr1_9985700_9985867 | 0.19 |
Ppp1r42 |
protein phosphatase 1, regulatory subunit 42 |
91 |
0.94 |
| chr7_71872986_71873347 | 0.19 |
4930441H08Rik |
RIKEN cDNA 4930441H08 gene |
36527 |
0.18 |
| chr10_87923075_87923294 | 0.19 |
Tyms-ps |
thymidylate synthase, pseudogene |
43663 |
0.12 |
| chr5_3386445_3386622 | 0.19 |
Cdk6 |
cyclin-dependent kinase 6 |
42221 |
0.13 |
| chr5_74316459_74316619 | 0.19 |
Gm15981 |
predicted gene 15981 |
495 |
0.77 |
| chr19_31905786_31905989 | 0.19 |
Gm19241 |
predicted gene, 19241 |
7757 |
0.22 |
| chr10_68179447_68179622 | 0.19 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
42908 |
0.17 |
| chr19_38399004_38399174 | 0.19 |
Slc35g1 |
solute carrier family 35, member G1 |
3048 |
0.21 |
| chr15_59055272_59055945 | 0.19 |
Mtss1 |
MTSS I-BAR domain containing 1 |
144 |
0.97 |
| chr5_143055727_143055894 | 0.19 |
Gm43378 |
predicted gene 43378 |
5767 |
0.15 |
| chrX_161292140_161292301 | 0.19 |
Gm15262 |
predicted gene 15262 |
4764 |
0.25 |
| chr4_124709849_124710395 | 0.18 |
Sf3a3 |
splicing factor 3a, subunit 3 |
4654 |
0.09 |
| chr17_26123354_26123516 | 0.18 |
Mrpl28 |
mitochondrial ribosomal protein L28 |
65 |
0.93 |
| chr7_141477112_141477277 | 0.18 |
Tspan4 |
tetraspanin 4 |
794 |
0.33 |
| chr6_125319120_125319346 | 0.18 |
Scnn1a |
sodium channel, nonvoltage-gated 1 alpha |
1426 |
0.25 |
| chr8_70212287_70212467 | 0.18 |
Slc25a42 |
solute carrier family 25, member 42 |
72 |
0.95 |
| chr5_90450366_90450538 | 0.18 |
Alb |
albumin |
10445 |
0.16 |
| chr7_132554752_132554903 | 0.18 |
Oat |
ornithine aminotransferase |
20891 |
0.15 |
| chr2_114274553_114274847 | 0.18 |
Gm29234 |
predicted gene 29234 |
1265 |
0.55 |
| chr1_13116391_13116585 | 0.18 |
Prdm14 |
PR domain containing 14 |
10675 |
0.15 |
| chr1_38449538_38449710 | 0.18 |
Gm34727 |
predicted gene, 34727 |
37895 |
0.18 |
| chr9_78513759_78514095 | 0.18 |
Gm47430 |
predicted gene, 47430 |
6619 |
0.13 |
| chr1_84056873_84057054 | 0.18 |
Pid1 |
phosphotyrosine interaction domain containing 1 |
3710 |
0.31 |
| chr11_119216234_119216406 | 0.18 |
Gm11752 |
predicted gene 11752 |
3840 |
0.14 |
| chr13_99238383_99238564 | 0.18 |
Zfp366 |
zinc finger protein 366 |
53650 |
0.12 |
| chr16_49887202_49887530 | 0.18 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
20533 |
0.26 |
| chr7_77787283_77787474 | 0.18 |
Gm23239 |
predicted gene, 23239 |
122942 |
0.06 |
| chr13_81633136_81633428 | 0.18 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
128 |
0.96 |
| chr4_55902839_55903035 | 0.18 |
Gm12519 |
predicted gene 12519 |
90802 |
0.09 |
| chr7_68687331_68687482 | 0.18 |
Gm44692 |
predicted gene 44692 |
39061 |
0.15 |
| chr7_99180189_99180656 | 0.18 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
129 |
0.94 |
| chr9_96223410_96223561 | 0.18 |
Gm37195 |
predicted gene, 37195 |
4355 |
0.2 |
| chr8_79288742_79288893 | 0.18 |
Mmaa |
methylmalonic aciduria (cobalamin deficiency) type A |
5863 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.2 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.2 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.0 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.0 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.1 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.0 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0004787 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |