Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
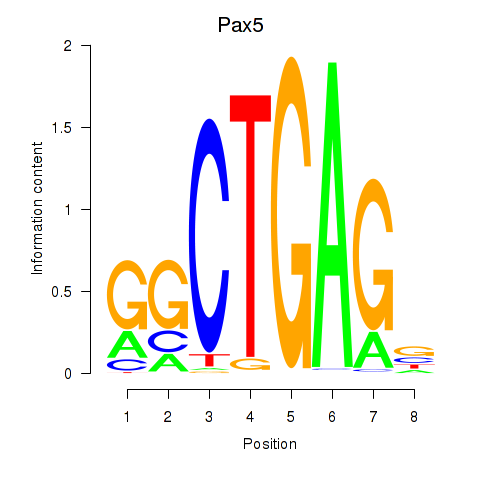
Results for Pax5
Z-value: 1.90
Transcription factors associated with Pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax5
|
ENSMUSG00000014030.9 | paired box 5 |
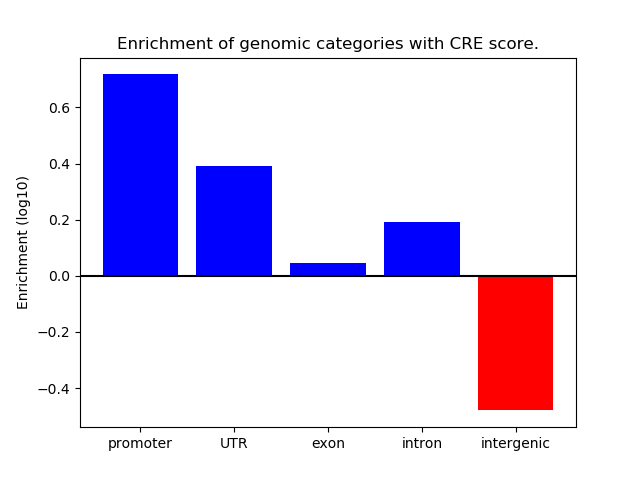
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_44645296_44645465 | Pax5 | 343 | 0.852879 | 0.67 | 1.5e-01 | Click! |
| chr4_44634038_44634207 | Pax5 | 11601 | 0.154084 | 0.23 | 6.7e-01 | Click! |
| chr4_44704491_44704674 | Pax5 | 576 | 0.644976 | 0.13 | 8.1e-01 | Click! |
| chr4_44633822_44633993 | Pax5 | 11816 | 0.153806 | 0.13 | 8.1e-01 | Click! |
Activity of the Pax5 motif across conditions
Conditions sorted by the z-value of the Pax5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
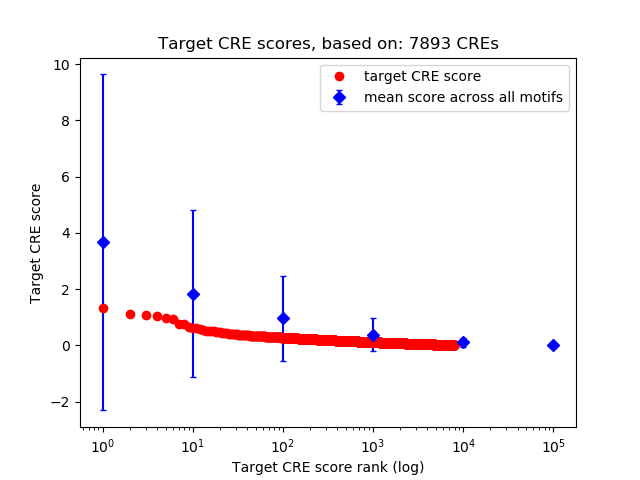
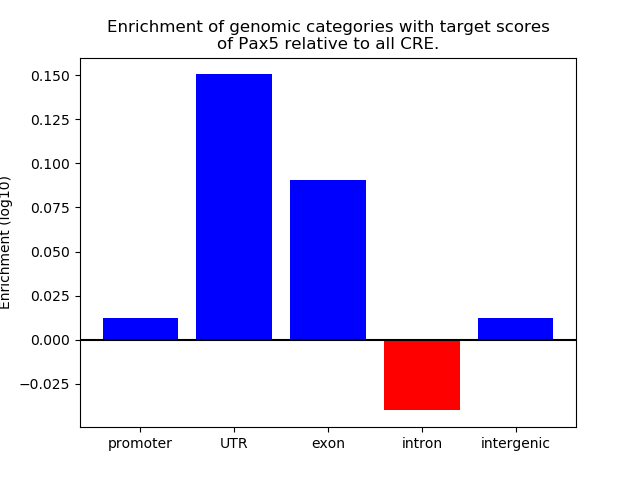
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_93959067_93959388 | 1.32 |
Gnao1 |
guanine nucleotide binding protein, alpha O |
3617 |
0.19 |
| chr11_110032371_110032522 | 1.10 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
2365 |
0.34 |
| chr11_110032912_110033079 | 1.06 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
2914 |
0.31 |
| chr19_46142576_46142826 | 1.06 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
1718 |
0.25 |
| chr19_36630127_36630294 | 0.98 |
Hectd2os |
Hectd2, opposite strand |
4186 |
0.26 |
| chr8_93958816_93959000 | 0.94 |
Gnao1 |
guanine nucleotide binding protein, alpha O |
3936 |
0.18 |
| chr8_10904318_10904469 | 0.77 |
Gm2814 |
predicted gene 2814 |
3835 |
0.12 |
| chr11_110032630_110032781 | 0.76 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
2624 |
0.32 |
| chr12_21165883_21166051 | 0.67 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
54013 |
0.1 |
| chr5_8961694_8961940 | 0.62 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
3265 |
0.17 |
| chr12_102683391_102683709 | 0.61 |
Mir1936 |
microRNA 1936 |
1470 |
0.28 |
| chr5_122530267_122530440 | 0.58 |
Gm22965 |
predicted gene, 22965 |
23564 |
0.09 |
| chr19_46134643_46134860 | 0.53 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
2538 |
0.17 |
| chr9_43673281_43673432 | 0.52 |
Gm5364 |
predicted gene 5364 |
9697 |
0.16 |
| chr3_41065898_41066144 | 0.52 |
Pgrmc2 |
progesterone receptor membrane component 2 |
4649 |
0.19 |
| chr8_93958496_93958784 | 0.52 |
Gnao1 |
guanine nucleotide binding protein, alpha O |
4204 |
0.18 |
| chr10_7976561_7976927 | 0.52 |
Tab2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
20514 |
0.19 |
| chr7_109115602_109115771 | 0.48 |
Gm44781 |
predicted gene 44781 |
27201 |
0.13 |
| chr7_12774535_12774865 | 0.48 |
Zscan18 |
zinc finger and SCAN domain containing 18 |
361 |
0.71 |
| chr10_24087275_24087474 | 0.46 |
Taar8b |
trace amine-associated receptor 8B |
4920 |
0.1 |
| chr7_44819003_44819183 | 0.45 |
Nup62 |
nucleoporin 62 |
656 |
0.42 |
| chr2_103846113_103846314 | 0.43 |
Gm13879 |
predicted gene 13879 |
2557 |
0.13 |
| chr15_102188714_102188935 | 0.43 |
Csad |
cysteine sulfinic acid decarboxylase |
150 |
0.92 |
| chr19_46141233_46141384 | 0.43 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
325 |
0.82 |
| chr19_42601924_42602199 | 0.42 |
Loxl4 |
lysyl oxidase-like 4 |
1260 |
0.45 |
| chr17_84896257_84896408 | 0.40 |
Gm49982 |
predicted gene, 49982 |
6237 |
0.19 |
| chr12_112559317_112559636 | 0.40 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
29308 |
0.12 |
| chr19_32534401_32535184 | 0.39 |
Gm36419 |
predicted gene, 36419 |
8556 |
0.18 |
| chr17_31902283_31902446 | 0.39 |
Gm50000 |
predicted gene, 50000 |
3186 |
0.18 |
| chr18_60662323_60662474 | 0.39 |
Synpo |
synaptopodin |
2256 |
0.28 |
| chr11_75430733_75430997 | 0.39 |
Gm12337 |
predicted gene 12337 |
3112 |
0.11 |
| chr2_166886612_166886763 | 0.38 |
Cse1l |
chromosome segregation 1-like (S. cerevisiae) |
19353 |
0.15 |
| chr11_101377034_101377689 | 0.37 |
G6pc |
glucose-6-phosphatase, catalytic |
9800 |
0.06 |
| chr6_99175134_99175460 | 0.37 |
Foxp1 |
forkhead box P1 |
12279 |
0.28 |
| chr13_93618635_93618791 | 0.36 |
Gm15622 |
predicted gene 15622 |
6669 |
0.17 |
| chr12_82856018_82856227 | 0.36 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
83033 |
0.09 |
| chr7_112208684_112208908 | 0.36 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
17060 |
0.24 |
| chr19_42461981_42462132 | 0.36 |
Crtac1 |
cartilage acidic protein 1 |
30271 |
0.15 |
| chr4_149777094_149777266 | 0.35 |
Gm16188 |
predicted gene 16188 |
928 |
0.36 |
| chr5_33220773_33221089 | 0.35 |
Spon2 |
spondin 2, extracellular matrix protein |
2476 |
0.24 |
| chr4_120092095_120092258 | 0.35 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
2313 |
0.34 |
| chr5_139871644_139871802 | 0.35 |
Gm42423 |
predicted gene 42423 |
24086 |
0.12 |
| chr18_20990195_20990346 | 0.35 |
Rnf138 |
ring finger protein 138 |
11071 |
0.2 |
| chr7_113239438_113239767 | 0.35 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
152 |
0.96 |
| chr10_96235051_96235242 | 0.34 |
4930459C07Rik |
RIKEN cDNA 4930459C07 gene |
8939 |
0.2 |
| chr15_67423581_67423756 | 0.34 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
196899 |
0.03 |
| chr17_5573057_5573246 | 0.34 |
Zdhhc14 |
zinc finger, DHHC domain containing 14 |
80594 |
0.08 |
| chr11_3984447_3984636 | 0.34 |
Gal3st1 |
galactose-3-O-sulfotransferase 1 |
828 |
0.43 |
| chr4_45850339_45850733 | 0.33 |
Stra6l |
STRA6-like |
1539 |
0.35 |
| chr10_8232552_8232849 | 0.33 |
Gm30906 |
predicted gene, 30906 |
56569 |
0.14 |
| chr15_10344412_10344563 | 0.33 |
Agxt2 |
alanine-glyoxylate aminotransferase 2 |
14045 |
0.18 |
| chr5_52982752_52983050 | 0.33 |
Gm30301 |
predicted gene, 30301 |
864 |
0.54 |
| chr18_38061485_38061666 | 0.33 |
Gm30093 |
predicted gene, 30093 |
38046 |
0.1 |
| chr9_22313296_22313633 | 0.33 |
Zfp810 |
zinc finger protein 810 |
5816 |
0.09 |
| chr8_12720450_12720739 | 0.32 |
Gm15348 |
predicted gene 15348 |
1467 |
0.36 |
| chr8_121584808_121584959 | 0.32 |
Gm17786 |
predicted gene, 17786 |
3118 |
0.14 |
| chr19_8872034_8872191 | 0.32 |
Ubxn1 |
UBX domain protein 1 |
452 |
0.5 |
| chr17_80833916_80834067 | 0.32 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
62802 |
0.11 |
| chr12_110865641_110866024 | 0.32 |
Mpc1-ps |
mitochondrial pyruvate carrier 1, pseudogene |
6938 |
0.09 |
| chr4_132974013_132974164 | 0.32 |
Fgr |
FGR proto-oncogene, Src family tyrosine kinase |
7 |
0.97 |
| chr19_21104697_21104848 | 0.32 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
62 |
0.98 |
| chr2_25472343_25472515 | 0.32 |
Ptgds |
prostaglandin D2 synthase (brain) |
2383 |
0.11 |
| chr12_110155051_110155218 | 0.31 |
3110009F21Rik |
RIKEN cDNA 3110009F21 gene |
2943 |
0.19 |
| chr7_120812600_120812786 | 0.31 |
Gm5737 |
predicted gene 5737 |
328 |
0.85 |
| chr3_93556679_93556843 | 0.31 |
S100a10 |
S100 calcium binding protein A10 (calpactin) |
1598 |
0.27 |
| chr8_119439834_119440348 | 0.31 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
2902 |
0.2 |
| chr4_149980982_149981133 | 0.31 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
20208 |
0.13 |
| chr15_38471890_38472111 | 0.31 |
G930009F23Rik |
RIKEN cDNA G930009F23 gene |
16866 |
0.13 |
| chr15_102185927_102186078 | 0.31 |
Csad |
cysteine sulfinic acid decarboxylase |
1597 |
0.23 |
| chr14_75178901_75179674 | 0.30 |
Lcp1 |
lymphocyte cytosolic protein 1 |
3079 |
0.21 |
| chr15_80684645_80684861 | 0.30 |
A430088P11Rik |
RIKEN cDNA A430088P11 gene |
12849 |
0.12 |
| chr9_43544933_43545084 | 0.30 |
Gm36855 |
predicted gene, 36855 |
24004 |
0.17 |
| chr6_145343998_145344149 | 0.30 |
Gm23498 |
predicted gene, 23498 |
22785 |
0.11 |
| chr4_148900818_148900969 | 0.30 |
Casz1 |
castor zinc finger 1 |
256 |
0.91 |
| chr9_65327502_65327653 | 0.30 |
Gm39363 |
predicted gene, 39363 |
4943 |
0.1 |
| chr1_93694081_93694251 | 0.29 |
Bok |
BCL2-related ovarian killer |
349 |
0.83 |
| chr1_120135102_120135272 | 0.29 |
3110009E18Rik |
RIKEN cDNA 3110009E18 gene |
13987 |
0.18 |
| chr10_108979737_108980117 | 0.29 |
Gm47477 |
predicted gene, 47477 |
23439 |
0.22 |
| chr12_77239645_77239931 | 0.29 |
Fut8 |
fucosyltransferase 8 |
723 |
0.56 |
| chr3_121279186_121279355 | 0.29 |
Gm30517 |
predicted gene, 30517 |
1746 |
0.27 |
| chr6_117298537_117298749 | 0.29 |
Rpl28-ps4 |
ribosomal protein L28, pseudogene 4 |
84577 |
0.08 |
| chr17_31401280_31401456 | 0.29 |
Pde9a |
phosphodiesterase 9A |
12788 |
0.13 |
| chr11_53883124_53883338 | 0.29 |
Slc22a5 |
solute carrier family 22 (organic cation transporter), member 5 |
8364 |
0.11 |
| chr11_116413032_116413183 | 0.29 |
Rnf157 |
ring finger protein 157 |
75 |
0.95 |
| chr18_57878443_57878604 | 0.29 |
Slc12a2 |
solute carrier family 12, member 2 |
155 |
0.97 |
| chr7_80653838_80654052 | 0.28 |
Gm15880 |
predicted gene 15880 |
17928 |
0.15 |
| chr4_155245715_155246079 | 0.28 |
Faap20 |
Fanconi anemia core complex associated protein 20 |
3905 |
0.19 |
| chr1_191924602_191924813 | 0.28 |
1700034H15Rik |
RIKEN cDNA 1700034H15 gene |
17180 |
0.13 |
| chr9_112103407_112103804 | 0.28 |
Mir128-2 |
microRNA 128-2 |
15106 |
0.26 |
| chr7_127809790_127809985 | 0.28 |
Stx1b |
syntaxin 1B |
1037 |
0.26 |
| chr3_98301677_98301831 | 0.28 |
Gm43189 |
predicted gene 43189 |
14766 |
0.14 |
| chr1_180216351_180216532 | 0.28 |
Gm37336 |
predicted gene, 37336 |
4040 |
0.17 |
| chr4_150012416_150012733 | 0.28 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
3551 |
0.18 |
| chr9_110702981_110703204 | 0.28 |
Ccdc12 |
coiled-coil domain containing 12 |
6802 |
0.12 |
| chr19_7122822_7123006 | 0.28 |
Flrt1 |
fibronectin leucine rich transmembrane protein 1 |
17185 |
0.12 |
| chr7_113214234_113214397 | 0.28 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
6758 |
0.23 |
| chr14_40924520_40924882 | 0.28 |
Tspan14 |
tetraspanin 14 |
9615 |
0.2 |
| chr16_23963940_23964091 | 0.28 |
Bcl6 |
B cell leukemia/lymphoma 6 |
8365 |
0.17 |
| chr3_98296908_98297093 | 0.28 |
Gm43189 |
predicted gene 43189 |
10012 |
0.15 |
| chr15_67337089_67337242 | 0.28 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
110396 |
0.07 |
| chr14_51009398_51009574 | 0.27 |
Rnase10 |
ribonuclease, RNase A family, 10 (non-active) |
1557 |
0.2 |
| chr2_154610076_154610298 | 0.27 |
Zfp341 |
zinc finger protein 341 |
3110 |
0.13 |
| chr19_47319894_47320142 | 0.27 |
Sh3pxd2a |
SH3 and PX domains 2A |
5267 |
0.2 |
| chr11_86916528_86916698 | 0.27 |
Ypel2 |
yippee like 2 |
55411 |
0.11 |
| chr17_28280441_28280673 | 0.27 |
Ppard |
peroxisome proliferator activator receptor delta |
8438 |
0.11 |
| chr12_3572503_3572654 | 0.27 |
Dtnb |
dystrobrevin, beta |
42 |
0.98 |
| chr14_34621069_34621237 | 0.27 |
Opn4 |
opsin 4 (melanopsin) |
21011 |
0.11 |
| chr8_46151778_46151954 | 0.27 |
Cfap97 |
cilia and flagella associated protein 97 |
95 |
0.57 |
| chr1_36536061_36536389 | 0.27 |
Ankrd23 |
ankyrin repeat domain 23 |
296 |
0.77 |
| chr8_120419577_120419740 | 0.27 |
Gm22715 |
predicted gene, 22715 |
23891 |
0.17 |
| chr17_56391344_56391495 | 0.27 |
Kdm4b |
lysine (K)-specific demethylase 4B |
2098 |
0.24 |
| chr4_118402815_118402985 | 0.27 |
Szt2 |
SZT2 subunit of KICSTOR complex |
6373 |
0.12 |
| chr5_8964820_8964997 | 0.27 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
6356 |
0.13 |
| chr15_100401091_100401280 | 0.27 |
Slc11a2 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
2058 |
0.2 |
| chr1_120111811_120112043 | 0.26 |
Dbi |
diazepam binding inhibitor |
8211 |
0.18 |
| chr1_120112071_120112794 | 0.26 |
Dbi |
diazepam binding inhibitor |
7706 |
0.18 |
| chr10_79706208_79706359 | 0.26 |
Bsg |
basigin |
1713 |
0.15 |
| chr14_26579689_26579840 | 0.26 |
Dennd6a |
DENN/MADD domain containing 6A |
186 |
0.91 |
| chr11_78418519_78418670 | 0.26 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
3623 |
0.12 |
| chr11_75433108_75433259 | 0.26 |
Gm12337 |
predicted gene 12337 |
5430 |
0.09 |
| chr10_43742623_43743061 | 0.26 |
Gm40634 |
predicted gene, 40634 |
245 |
0.89 |
| chr16_94536454_94536803 | 0.26 |
Vps26c |
VPS26 endosomal protein sorting factor C |
9798 |
0.18 |
| chr19_47369089_47369248 | 0.26 |
Sh3pxd2a |
SH3 and PX domains 2A |
41191 |
0.14 |
| chr11_112217336_112217487 | 0.26 |
Gm11680 |
predicted gene 11680 |
109973 |
0.07 |
| chr9_3199735_3199905 | 0.26 |
4930433N12Rik |
RIKEN cDNA 4930433N12 gene |
21 |
0.54 |
| chr12_21166146_21166319 | 0.26 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
54278 |
0.1 |
| chr1_72172049_72172205 | 0.26 |
Mreg |
melanoregulin |
40180 |
0.1 |
| chr1_191950425_191950576 | 0.26 |
Rd3 |
retinal degeneration 3 |
26870 |
0.1 |
| chr5_123228121_123228280 | 0.26 |
Psmd9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
10 |
0.96 |
| chr8_3407162_3407313 | 0.26 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
14164 |
0.15 |
| chr16_8566051_8566267 | 0.25 |
Abat |
4-aminobutyrate aminotransferase |
6743 |
0.17 |
| chr6_72604832_72604993 | 0.25 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
539 |
0.39 |
| chr3_89715420_89715571 | 0.25 |
Adar |
adenosine deaminase, RNA-specific |
163 |
0.94 |
| chr7_80628977_80629128 | 0.25 |
Crtc3 |
CREB regulated transcription coactivator 3 |
574 |
0.71 |
| chr10_41567217_41567368 | 0.25 |
5730435O14Rik |
RIKEN cDNA 5730435O14 gene |
724 |
0.57 |
| chr11_112217513_112217697 | 0.25 |
Gm11680 |
predicted gene 11680 |
110167 |
0.07 |
| chr15_76575014_76575219 | 0.25 |
Adck5 |
aarF domain containing kinase 5 |
1242 |
0.21 |
| chr10_80824337_80824490 | 0.25 |
Oaz1 |
ornithine decarboxylase antizyme 1 |
2243 |
0.12 |
| chr2_153528144_153528295 | 0.25 |
Nol4l |
nucleolar protein 4-like |
1752 |
0.37 |
| chr12_106468407_106468693 | 0.25 |
Gm3191 |
predicted gene 3191 |
9400 |
0.19 |
| chr3_14871524_14871675 | 0.25 |
Car3 |
carbonic anhydrase 3 |
2421 |
0.28 |
| chr4_124630137_124630316 | 0.25 |
Pou3f1 |
POU domain, class 3, transcription factor 1 |
26581 |
0.11 |
| chr9_48758222_48758532 | 0.24 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
77568 |
0.09 |
| chr9_108806919_108807090 | 0.24 |
Nckipsd |
NCK interacting protein with SH3 domain |
1364 |
0.23 |
| chr7_3331834_3332020 | 0.24 |
Cacng7 |
calcium channel, voltage-dependent, gamma subunit 7 |
1028 |
0.31 |
| chr19_47315149_47315552 | 0.24 |
Sh3pxd2a |
SH3 and PX domains 2A |
599 |
0.73 |
| chr18_44634075_44634320 | 0.24 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
27468 |
0.19 |
| chr14_48509565_48509731 | 0.24 |
Tmem260 |
transmembrane protein 260 |
3662 |
0.16 |
| chr4_35100180_35100588 | 0.24 |
Ifnk |
interferon kappa |
51672 |
0.12 |
| chr8_126754584_126754786 | 0.24 |
Gm45805 |
predicted gene 45805 |
3649 |
0.31 |
| chr19_61048830_61049007 | 0.24 |
Gm22520 |
predicted gene, 22520 |
35373 |
0.14 |
| chr4_144930054_144930220 | 0.24 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
11258 |
0.19 |
| chr11_95786990_95787141 | 0.24 |
Polr2k-ps |
polymerase (RNA) II (DNA directed) polypeptide K, pseudogene |
25574 |
0.1 |
| chr8_126586607_126587402 | 0.24 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
6982 |
0.23 |
| chr9_57281733_57281969 | 0.24 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
19239 |
0.14 |
| chr1_72810094_72810265 | 0.24 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
14324 |
0.21 |
| chr17_29078234_29078394 | 0.24 |
Trp53cor1 |
tumor protein p53 pathway corepressor 1 |
865 |
0.39 |
| chr4_119223756_119224101 | 0.23 |
Gm12898 |
predicted gene 12898 |
754 |
0.42 |
| chr1_74119208_74119359 | 0.23 |
Tns1 |
tensin 1 |
5164 |
0.16 |
| chr16_4029500_4029663 | 0.23 |
Dnase1 |
deoxyribonuclease I |
7361 |
0.1 |
| chr17_23675280_23675431 | 0.23 |
Tnfrsf12a |
tumor necrosis factor receptor superfamily, member 12a |
1198 |
0.16 |
| chr3_132815131_132815356 | 0.23 |
Gm42876 |
predicted gene 42876 |
8990 |
0.14 |
| chr9_119145447_119145657 | 0.23 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
4511 |
0.13 |
| chr6_72600962_72601192 | 0.23 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
2449 |
0.13 |
| chr5_139338482_139338648 | 0.23 |
Cox19 |
cytochrome c oxidase assembly protein 19 |
1010 |
0.41 |
| chr11_51825017_51825309 | 0.23 |
Jade2 |
jade family PHD finger 2 |
31962 |
0.13 |
| chr4_119225130_119225281 | 0.23 |
Gm12898 |
predicted gene 12898 |
2031 |
0.15 |
| chr17_31386878_31387227 | 0.23 |
Pde9a |
phosphodiesterase 9A |
758 |
0.59 |
| chr2_167877367_167877523 | 0.23 |
Gm14319 |
predicted gene 14319 |
18860 |
0.16 |
| chr9_57785755_57786023 | 0.23 |
Arid3b |
AT rich interactive domain 3B (BRIGHT-like) |
5759 |
0.16 |
| chr10_75044109_75044374 | 0.23 |
Rab36 |
RAB36, member RAS oncogene family |
6018 |
0.16 |
| chr19_23034228_23034427 | 0.23 |
Gm50136 |
predicted gene, 50136 |
27127 |
0.18 |
| chr10_13869659_13869820 | 0.23 |
Aig1 |
androgen-induced 1 |
759 |
0.5 |
| chr10_17726393_17726767 | 0.23 |
Cited2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
3338 |
0.27 |
| chr4_151179063_151179232 | 0.23 |
Camta1 |
calmodulin binding transcription activator 1 |
39563 |
0.17 |
| chr2_158095379_158095534 | 0.23 |
2010009K17Rik |
RIKEN cDNA 2010009K17 gene |
3659 |
0.19 |
| chr13_58217381_58217535 | 0.23 |
Gm48432 |
predicted gene, 48432 |
1261 |
0.27 |
| chr4_115518540_115518728 | 0.23 |
Cyp4a10 |
cytochrome P450, family 4, subfamily a, polypeptide 10 |
346 |
0.81 |
| chr9_110246098_110246279 | 0.23 |
Cspg5 |
chondroitin sulfate proteoglycan 5 |
123 |
0.94 |
| chr11_62495246_62495444 | 0.23 |
Gm12278 |
predicted gene 12278 |
12548 |
0.1 |
| chr14_31660786_31660937 | 0.23 |
Hacl1 |
2-hydroxyacyl-CoA lyase 1 |
19575 |
0.13 |
| chr13_60508060_60508255 | 0.23 |
A530001N23Rik |
RIKEN cDNA A530001N23 gene |
19349 |
0.15 |
| chr11_5064664_5064914 | 0.23 |
Gas2l1 |
growth arrest-specific 2 like 1 |
62 |
0.95 |
| chr11_3509541_3509989 | 0.23 |
Selenom |
selenoprotein M |
4919 |
0.1 |
| chr11_4883931_4884115 | 0.23 |
Nipsnap1 |
nipsnap homolog 1 |
759 |
0.54 |
| chr6_54719950_54720402 | 0.23 |
Gm44008 |
predicted gene, 44008 |
26530 |
0.15 |
| chr11_112219735_112219893 | 0.23 |
Gm11680 |
predicted gene 11680 |
112376 |
0.07 |
| chr4_41638036_41638217 | 0.23 |
Gm12406 |
predicted gene 12406 |
1252 |
0.26 |
| chr17_33555577_33555756 | 0.23 |
Myo1f |
myosin IF |
41 |
0.97 |
| chr2_179292597_179292961 | 0.22 |
Gm14293 |
predicted gene 14293 |
52289 |
0.14 |
| chr19_61080476_61080639 | 0.22 |
Zfp950 |
zinc finger protein 950 |
36816 |
0.13 |
| chr3_98302260_98302728 | 0.22 |
Gm43189 |
predicted gene 43189 |
15506 |
0.14 |
| chr1_191417042_191417418 | 0.22 |
Gm2272 |
predicted gene 2272 |
9564 |
0.15 |
| chr10_128586195_128586346 | 0.22 |
Mir6917 |
microRNA 6917 |
1773 |
0.14 |
| chr8_71476414_71476589 | 0.22 |
Dda1 |
DET1 and DDB1 associated 1 |
6090 |
0.08 |
| chr15_97075953_97076132 | 0.22 |
Slc38a4 |
solute carrier family 38, member 4 |
20086 |
0.24 |
| chr10_59784742_59784904 | 0.22 |
Gm17059 |
predicted gene 17059 |
15431 |
0.14 |
| chr7_24598283_24598438 | 0.22 |
Zfp575 |
zinc finger protein 575 |
10719 |
0.08 |
| chr15_31409939_31410113 | 0.22 |
Gm6361 |
predicted gene 6361 |
36791 |
0.09 |
| chrX_94122808_94122959 | 0.22 |
Zfx |
zinc finger protein X-linked |
17 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.2 | 0.5 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.3 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.3 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.2 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.1 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.2 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.1 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.2 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.2 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) |
| 0.0 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0006113 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.2 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:1902990 | telomere maintenance via semi-conservative replication(GO:0032201) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.4 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.0 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.0 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.0 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:2001225 | regulation of chloride transport(GO:2001225) |
| 0.0 | 0.1 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.0 | GO:0072197 | ureter morphogenesis(GO:0072197) |
| 0.0 | 0.0 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.0 | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.0 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0032762 | mast cell cytokine production(GO:0032762) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.5 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.0 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.0 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.0 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.0 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.0 | GO:0071899 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:1903393 | positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.0 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.0 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.1 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0071420 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.0 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.0 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.0 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.0 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.0 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0018657 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 1.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.0 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.0 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |