Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
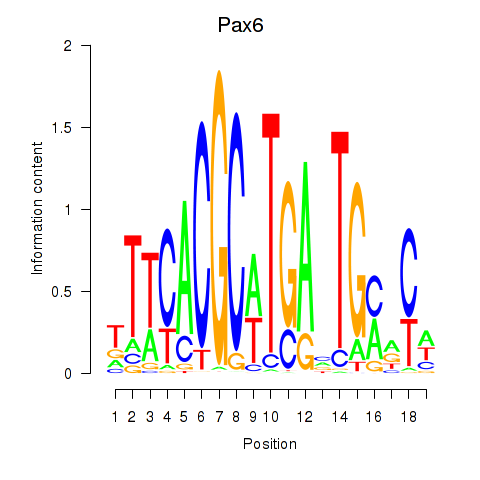
Results for Pax6
Z-value: 0.81
Transcription factors associated with Pax6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax6
|
ENSMUSG00000027168.15 | paired box 6 |
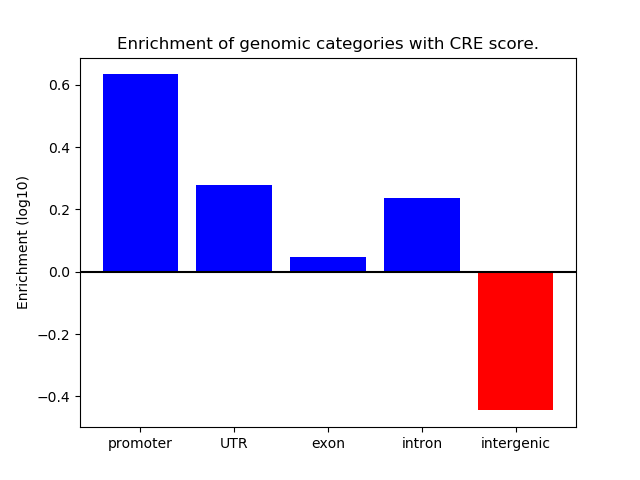
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_105675376_105675546 | Pax6 | 28 | 0.972119 | 0.37 | 4.7e-01 | Click! |
| chr2_105673831_105674008 | Pax6 | 44 | 0.969838 | -0.19 | 7.1e-01 | Click! |
| chr2_105687448_105687763 | Pax6 | 2283 | 0.259778 | -0.10 | 8.6e-01 | Click! |
| chr2_105674236_105674387 | Pax6 | 348 | 0.850363 | -0.03 | 9.5e-01 | Click! |
Activity of the Pax6 motif across conditions
Conditions sorted by the z-value of the Pax6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
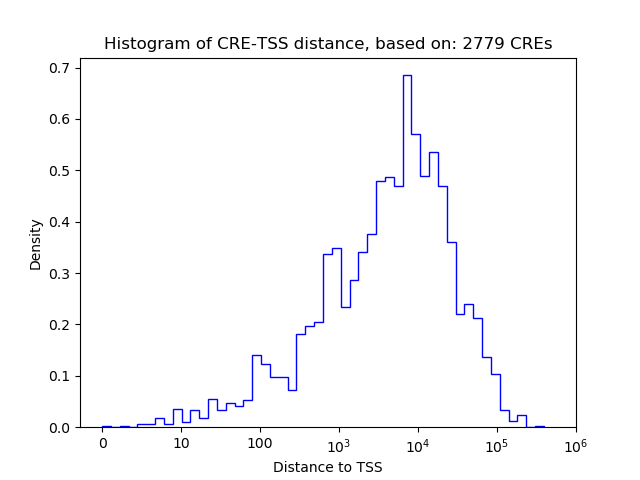
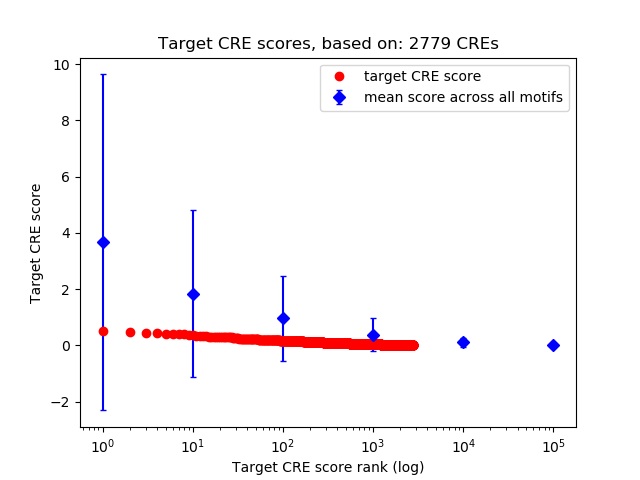
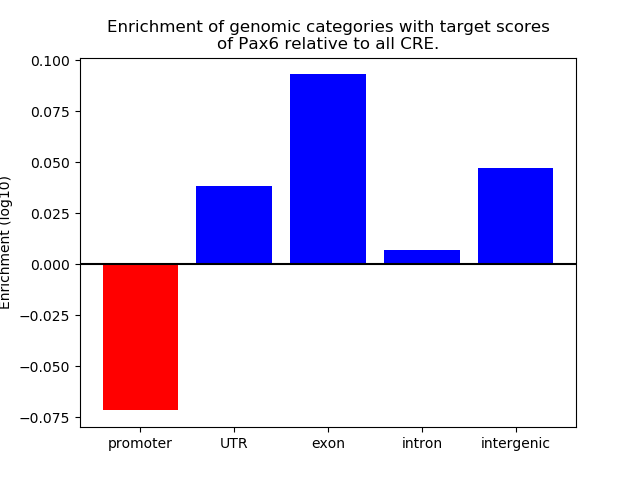
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_74331641_74331792 | 0.51 |
Pnkd |
paroxysmal nonkinesiogenic dyskinesia |
894 |
0.38 |
| chr4_34495112_34495276 | 0.46 |
Gm12350 |
predicted gene 12350 |
9362 |
0.19 |
| chr5_122263936_122264091 | 0.44 |
Tctn1 |
tectonic family member 1 |
402 |
0.76 |
| chr12_21183267_21183628 | 0.44 |
AC156032.1 |
|
63876 |
0.08 |
| chr1_180161827_180161978 | 0.41 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
895 |
0.53 |
| chr9_65333970_65334121 | 0.40 |
Gm39363 |
predicted gene, 39363 |
1525 |
0.2 |
| chr9_53772188_53772357 | 0.39 |
Slc35f2 |
solute carrier family 35, member F2 |
734 |
0.65 |
| chr15_58701542_58701739 | 0.39 |
Fer1l6 |
fer-1-like 6 (C. elegans) |
53135 |
0.12 |
| chr10_96235051_96235242 | 0.38 |
4930459C07Rik |
RIKEN cDNA 4930459C07 gene |
8939 |
0.2 |
| chr17_45322913_45323086 | 0.36 |
Cdc5l |
cell division cycle 5-like (S. pombe) |
92685 |
0.06 |
| chr15_67062566_67062733 | 0.34 |
Gm31342 |
predicted gene, 31342 |
22591 |
0.2 |
| chr13_30235169_30235331 | 0.34 |
Mboat1 |
membrane bound O-acyltransferase domain containing 1 |
3397 |
0.27 |
| chr17_30609874_30610226 | 0.32 |
Glo1 |
glyoxalase 1 |
2519 |
0.15 |
| chr19_38356216_38356572 | 0.31 |
Gm50150 |
predicted gene, 50150 |
13780 |
0.14 |
| chr2_129380377_129380528 | 0.31 |
Spcs2-ps |
signal peptidase complex subunit 2, pseudogene |
1056 |
0.39 |
| chr8_45295376_45295693 | 0.30 |
Klkb1 |
kallikrein B, plasma 1 |
675 |
0.64 |
| chr8_10939586_10939983 | 0.30 |
Gm44955 |
predicted gene 44955 |
8106 |
0.11 |
| chr19_34518293_34518456 | 0.30 |
Lipa |
lysosomal acid lipase A |
7394 |
0.14 |
| chr9_95571722_95572131 | 0.30 |
Paqr9 |
progestin and adipoQ receptor family member IX |
12269 |
0.13 |
| chr18_53135544_53135717 | 0.30 |
Snx2 |
sorting nexin 2 |
40686 |
0.18 |
| chr19_21824245_21824555 | 0.30 |
Gm50130 |
predicted gene, 50130 |
9356 |
0.22 |
| chr10_84055045_84055203 | 0.30 |
Gm37908 |
predicted gene, 37908 |
6634 |
0.2 |
| chr2_79254721_79254886 | 0.29 |
Itga4 |
integrin alpha 4 |
623 |
0.79 |
| chr6_121870418_121870617 | 0.28 |
Mug1 |
murinoglobulin 1 |
15060 |
0.19 |
| chr11_97884607_97884760 | 0.28 |
Gm11630 |
predicted gene 11630 |
89 |
0.87 |
| chr5_120125413_120125579 | 0.28 |
Rbm19 |
RNA binding motif protein 19 |
6900 |
0.2 |
| chr12_25092894_25093391 | 0.28 |
Id2 |
inhibitor of DNA binding 2 |
2945 |
0.22 |
| chr7_130716830_130716981 | 0.28 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
3228 |
0.24 |
| chr6_90763367_90763528 | 0.27 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
694 |
0.67 |
| chr12_80136272_80136487 | 0.26 |
2310015A10Rik |
RIKEN cDNA 2310015A10 gene |
3535 |
0.16 |
| chr12_102683391_102683709 | 0.24 |
Mir1936 |
microRNA 1936 |
1470 |
0.28 |
| chr9_63613110_63613389 | 0.24 |
Aagab |
alpha- and gamma-adaptin binding protein |
3739 |
0.23 |
| chr15_89475186_89475337 | 0.24 |
Arsa |
arylsulfatase A |
852 |
0.31 |
| chr4_15091382_15091540 | 0.24 |
Necab1 |
N-terminal EF-hand calcium binding protein 1 |
57500 |
0.12 |
| chr4_129795439_129795602 | 0.24 |
Ptp4a2 |
protein tyrosine phosphatase 4a2 |
15699 |
0.12 |
| chr5_127633759_127633922 | 0.23 |
Slc15a4 |
solute carrier family 15, member 4 |
943 |
0.47 |
| chr1_136142886_136143042 | 0.22 |
Kif21b |
kinesin family member 21B |
11510 |
0.11 |
| chr6_60600291_60600442 | 0.22 |
Gm35386 |
predicted gene, 35386 |
13588 |
0.26 |
| chr11_51833074_51833257 | 0.22 |
Jade2 |
jade family PHD finger 2 |
23960 |
0.15 |
| chr13_95952461_95952612 | 0.22 |
Sv2c |
synaptic vesicle glycoprotein 2c |
32314 |
0.16 |
| chr1_133825700_133825858 | 0.22 |
Gm10537 |
predicted gene 10537 |
3749 |
0.17 |
| chrX_48193537_48193988 | 0.22 |
Zdhhc9 |
zinc finger, DHHC domain containing 9 |
14566 |
0.16 |
| chr2_114835914_114836098 | 0.22 |
Gm13974 |
predicted gene 13974 |
18087 |
0.25 |
| chr5_112292069_112292220 | 0.22 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
3382 |
0.15 |
| chr2_84184780_84185142 | 0.22 |
Gm25972 |
predicted gene, 25972 |
34906 |
0.15 |
| chr3_83002849_83003457 | 0.22 |
Fgg |
fibrinogen gamma chain |
4571 |
0.17 |
| chr5_52975314_52975474 | 0.22 |
Gm30301 |
predicted gene, 30301 |
6643 |
0.16 |
| chr1_184290067_184290274 | 0.22 |
Gm37223 |
predicted gene, 37223 |
68159 |
0.11 |
| chr3_137864426_137864597 | 0.21 |
H2az1 |
H2A.Z variant histone 1 |
24 |
0.62 |
| chr16_16224499_16224674 | 0.21 |
Pkp2 |
plakophilin 2 |
11268 |
0.19 |
| chr5_100872116_100872267 | 0.21 |
Gm42987 |
predicted gene 42987 |
11942 |
0.11 |
| chr1_180813964_180814133 | 0.21 |
H3f3a |
H3.3 histone A |
105 |
0.7 |
| chr9_95573382_95573533 | 0.21 |
Paqr9 |
progestin and adipoQ receptor family member IX |
13800 |
0.12 |
| chr15_62205085_62205531 | 0.21 |
Pvt1 |
Pvt1 oncogene |
12915 |
0.24 |
| chr11_30792834_30792985 | 0.20 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
18628 |
0.14 |
| chr10_19991939_19992101 | 0.20 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
40965 |
0.16 |
| chr16_16225035_16225410 | 0.20 |
Pkp2 |
plakophilin 2 |
11904 |
0.19 |
| chr18_46716574_46716962 | 0.20 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
11261 |
0.13 |
| chr6_94549050_94549217 | 0.20 |
Slc25a26 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
14993 |
0.2 |
| chr13_36459438_36459830 | 0.20 |
Gm48763 |
predicted gene, 48763 |
354 |
0.9 |
| chr14_17894891_17895062 | 0.20 |
Thrb |
thyroid hormone receptor beta |
67845 |
0.12 |
| chr10_127289815_127290011 | 0.20 |
Gm4189 |
predicted gene 4189 |
811 |
0.21 |
| chr12_83375040_83375279 | 0.20 |
Dpf3 |
D4, zinc and double PHD fingers, family 3 |
24264 |
0.22 |
| chr2_167574964_167575153 | 0.19 |
Gm11475 |
predicted gene 11475 |
16337 |
0.11 |
| chr2_152792514_152792820 | 0.19 |
Gm23802 |
predicted gene, 23802 |
19778 |
0.11 |
| chr1_172312301_172312452 | 0.19 |
Igsf8 |
immunoglobulin superfamily, member 8 |
9 |
0.96 |
| chr19_47511131_47511282 | 0.19 |
Gm19557 |
predicted gene, 19557 |
1776 |
0.29 |
| chr12_12320785_12320940 | 0.19 |
Fam49a |
family with sequence similarity 49, member A |
58673 |
0.14 |
| chr19_12634142_12634556 | 0.19 |
Glyat |
glycine-N-acyltransferase |
816 |
0.44 |
| chr12_75694445_75694598 | 0.19 |
Wdr89 |
WD repeat domain 89 |
24984 |
0.18 |
| chr5_8849350_8849972 | 0.19 |
Abcb1b |
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
328 |
0.86 |
| chr2_163480327_163480478 | 0.19 |
Fitm2 |
fat storage-inducing transmembrane protein 2 |
7773 |
0.11 |
| chr10_118737027_118737347 | 0.19 |
Gm47425 |
predicted gene, 47425 |
17759 |
0.17 |
| chr7_25623384_25623543 | 0.18 |
Dmac2 |
distal membrane arm assembly complex 2 |
2560 |
0.12 |
| chr7_130789536_130789700 | 0.18 |
Btbd16 |
BTB (POZ) domain containing 16 |
15451 |
0.14 |
| chr9_63201496_63201967 | 0.18 |
Skor1 |
SKI family transcriptional corepressor 1 |
52770 |
0.12 |
| chr4_33243089_33243244 | 0.18 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
4409 |
0.19 |
| chr19_41346470_41346772 | 0.18 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
38475 |
0.17 |
| chr1_182609752_182609909 | 0.18 |
Capn8 |
calpain 8 |
44792 |
0.12 |
| chr1_86394431_86394582 | 0.18 |
Nmur1 |
neuromedin U receptor 1 |
3684 |
0.12 |
| chr15_99724752_99724954 | 0.18 |
Gm16537 |
predicted gene 16537 |
379 |
0.58 |
| chr4_150334328_150334517 | 0.18 |
Gm23209 |
predicted gene, 23209 |
13229 |
0.17 |
| chr17_13408343_13408512 | 0.18 |
Gm8597 |
predicted gene 8597 |
4236 |
0.13 |
| chr9_64049525_64049685 | 0.18 |
Gm25606 |
predicted gene, 25606 |
1109 |
0.4 |
| chr2_158225371_158225751 | 0.18 |
D630003M21Rik |
RIKEN cDNA D630003M21 gene |
3661 |
0.17 |
| chr2_119355713_119355881 | 0.17 |
Chac1 |
ChaC, cation transport regulator 1 |
4568 |
0.16 |
| chr13_32828221_32828372 | 0.17 |
Wrnip1 |
Werner helicase interacting protein 1 |
7530 |
0.14 |
| chr6_124901137_124901288 | 0.17 |
Lag3 |
lymphocyte-activation gene 3 |
8839 |
0.07 |
| chr17_84896467_84896802 | 0.17 |
Gm49982 |
predicted gene, 49982 |
5935 |
0.19 |
| chr16_24223468_24223662 | 0.17 |
Gm31814 |
predicted gene, 31814 |
7041 |
0.23 |
| chr1_58718073_58718224 | 0.17 |
Cflar |
CASP8 and FADD-like apoptosis regulator |
4262 |
0.15 |
| chr10_71328527_71328693 | 0.17 |
Rpl19-ps2 |
ribosomal protein L19, pseudogene 2 |
9844 |
0.12 |
| chr19_36796940_36797118 | 0.17 |
Gm50112 |
predicted gene, 50112 |
21619 |
0.18 |
| chr10_77051818_77052096 | 0.17 |
Col18a1 |
collagen, type XVIII, alpha 1 |
8040 |
0.14 |
| chr8_93333023_93333204 | 0.17 |
Ces1g |
carboxylesterase 1G |
4195 |
0.17 |
| chr4_104822600_104822778 | 0.17 |
C8b |
complement component 8, beta polypeptide |
23481 |
0.18 |
| chr11_58884795_58884971 | 0.17 |
Gm12256 |
predicted gene 12256 |
4827 |
0.08 |
| chr12_82397673_82398162 | 0.17 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
22698 |
0.24 |
| chr5_115528254_115528453 | 0.17 |
Pxn |
paxillin |
8048 |
0.09 |
| chr1_62425224_62425405 | 0.17 |
Pard3bos3 |
par-3 family cell polarity regulator beta, opposite strand 3 |
73947 |
0.11 |
| chr6_128519043_128519655 | 0.17 |
Pzp |
PZP, alpha-2-macroglobulin like |
7354 |
0.09 |
| chr12_69215107_69215258 | 0.16 |
Pole2 |
polymerase (DNA directed), epsilon 2 (p59 subunit) |
1281 |
0.21 |
| chr1_179961858_179962048 | 0.16 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
843 |
0.65 |
| chr13_93618437_93618616 | 0.16 |
Gm15622 |
predicted gene 15622 |
6856 |
0.17 |
| chr19_42562288_42562441 | 0.16 |
Gm16541 |
predicted gene 16541 |
731 |
0.61 |
| chr2_68001837_68002008 | 0.16 |
Gm13607 |
predicted gene 13607 |
79869 |
0.09 |
| chr5_102511749_102511901 | 0.16 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
28536 |
0.2 |
| chr6_108507437_108507634 | 0.16 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
778 |
0.59 |
| chr2_90474865_90475228 | 0.16 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
3710 |
0.21 |
| chr4_35134987_35135178 | 0.16 |
Ifnk |
interferon kappa |
16974 |
0.18 |
| chr1_88630811_88630966 | 0.16 |
Gm38130 |
predicted gene, 38130 |
22197 |
0.15 |
| chr5_65350576_65350727 | 0.16 |
Klb |
klotho beta |
2243 |
0.19 |
| chr2_35215153_35215312 | 0.16 |
Gm13605 |
predicted gene 13605 |
9301 |
0.15 |
| chr14_122464092_122464278 | 0.16 |
Zic5 |
zinc finger protein of the cerebellum 5 |
1492 |
0.27 |
| chr17_80479277_80479673 | 0.16 |
Sos1 |
SOS Ras/Rac guanine nucleotide exchange factor 1 |
494 |
0.83 |
| chr4_57954096_57954315 | 0.16 |
Txn1 |
thioredoxin 1 |
2206 |
0.32 |
| chr14_18221191_18221360 | 0.16 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
8903 |
0.14 |
| chr14_21096961_21097138 | 0.16 |
Adk |
adenosine kinase |
20897 |
0.2 |
| chr6_48569197_48569793 | 0.16 |
Rarres2 |
retinoic acid receptor responder (tazarotene induced) 2 |
3151 |
0.09 |
| chr3_88102838_88102989 | 0.15 |
Iqgap3 |
IQ motif containing GTPase activating protein 3 |
2229 |
0.18 |
| chr8_72665741_72665896 | 0.15 |
Nwd1 |
NACHT and WD repeat domain containing 1 |
8732 |
0.15 |
| chr11_106579550_106579738 | 0.15 |
Tex2 |
testis expressed gene 2 |
531 |
0.78 |
| chr12_25416673_25416856 | 0.15 |
Gm36723 |
predicted gene, 36723 |
22009 |
0.24 |
| chr5_21053272_21053655 | 0.15 |
Ptpn12 |
protein tyrosine phosphatase, non-receptor type 12 |
2262 |
0.26 |
| chr5_36678947_36679110 | 0.15 |
Gm25767 |
predicted gene, 25767 |
7370 |
0.14 |
| chr14_20208430_20208743 | 0.15 |
Gm48395 |
predicted gene, 48395 |
6916 |
0.12 |
| chr1_106200351_106200637 | 0.15 |
Gm38235 |
predicted gene, 38235 |
18820 |
0.17 |
| chr2_29739509_29739665 | 0.15 |
Gm9823 |
predicted gene 9823 |
922 |
0.43 |
| chr9_65831812_65832370 | 0.15 |
n-R5s85 |
nuclear encoded rRNA 5S 85 |
4262 |
0.15 |
| chr7_46827705_46827889 | 0.15 |
Gm45308 |
predicted gene 45308 |
4667 |
0.1 |
| chr3_108011690_108011861 | 0.15 |
Gstm1 |
glutathione S-transferase, mu 1 |
1541 |
0.17 |
| chr6_141944108_141944325 | 0.15 |
Slco1a1 |
solute carrier organic anion transporter family, member 1a1 |
2594 |
0.32 |
| chr11_49085302_49085517 | 0.15 |
Ifi47 |
interferon gamma inducible protein 47 |
1291 |
0.22 |
| chr11_84959738_84959922 | 0.15 |
Car4 |
carbonic anhydrase 4 |
1726 |
0.3 |
| chr7_49233095_49233247 | 0.15 |
Nav2 |
neuron navigator 2 |
13018 |
0.21 |
| chr11_120280389_120280554 | 0.15 |
Gm47297 |
predicted gene, 47297 |
5984 |
0.11 |
| chr13_54566023_54566188 | 0.15 |
4833439L19Rik |
RIKEN cDNA 4833439L19 gene |
670 |
0.53 |
| chr2_153600154_153600312 | 0.15 |
Commd7 |
COMM domain containing 7 |
32498 |
0.13 |
| chr17_29486635_29486945 | 0.15 |
Pim1 |
proviral integration site 1 |
3963 |
0.13 |
| chr19_7090027_7090210 | 0.15 |
AC109619.1 |
novel transcript, antisense to Macrod1 |
1697 |
0.24 |
| chr3_38259341_38259510 | 0.15 |
Gm2965 |
predicted gene 2965 |
35974 |
0.16 |
| chr13_23730927_23731223 | 0.14 |
Gm11338 |
predicted gene 11338 |
2527 |
0.07 |
| chr2_73455695_73455846 | 0.14 |
Wipf1 |
WAS/WASL interacting protein family, member 1 |
1791 |
0.28 |
| chr12_80113015_80113206 | 0.14 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
97 |
0.95 |
| chr4_137747156_137747355 | 0.14 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
108 |
0.97 |
| chr17_31386878_31387227 | 0.14 |
Pde9a |
phosphodiesterase 9A |
758 |
0.59 |
| chr16_92433602_92433820 | 0.14 |
Gm46555 |
predicted gene, 46555 |
23766 |
0.12 |
| chr12_113255450_113255616 | 0.14 |
Gm25622 |
predicted gene, 25622 |
41384 |
0.09 |
| chr2_164769333_164769540 | 0.14 |
Gm11457 |
predicted gene 11457 |
183 |
0.71 |
| chr13_39657075_39657270 | 0.14 |
Gm47352 |
predicted gene, 47352 |
7661 |
0.21 |
| chr18_75821165_75821446 | 0.14 |
Zbtb7c |
zinc finger and BTB domain containing 7C |
1090 |
0.57 |
| chr15_67345700_67345857 | 0.14 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
119009 |
0.06 |
| chr5_93250297_93250457 | 0.14 |
Ccng2 |
cyclin G2 |
16880 |
0.14 |
| chr5_64344454_64344620 | 0.14 |
Gm43838 |
predicted gene 43838 |
15977 |
0.14 |
| chr4_70410468_70410632 | 0.14 |
Cdk5rap2 |
CDK5 regulatory subunit associated protein 2 |
107 |
0.98 |
| chr16_24090719_24090960 | 0.14 |
Gm31583 |
predicted gene, 31583 |
750 |
0.64 |
| chr2_84386945_84387393 | 0.14 |
Calcrl |
calcitonin receptor-like |
11811 |
0.2 |
| chr4_57295426_57295990 | 0.14 |
Gm12536 |
predicted gene 12536 |
4388 |
0.21 |
| chr10_8873465_8873676 | 0.14 |
Gm26674 |
predicted gene, 26674 |
11067 |
0.15 |
| chr11_53783385_53783552 | 0.14 |
Gm17334 |
predicted gene, 17334 |
10273 |
0.11 |
| chr12_79561056_79561219 | 0.14 |
Rad51b |
RAD51 paralog B |
233784 |
0.02 |
| chr15_76232589_76232802 | 0.14 |
Plec |
plectin |
121 |
0.87 |
| chr19_4698221_4698442 | 0.14 |
Gm960 |
predicted gene 960 |
16 |
0.96 |
| chr15_28057589_28057753 | 0.14 |
Gm19883 |
predicted gene, 19883 |
16662 |
0.18 |
| chr6_5509839_5510005 | 0.14 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
13613 |
0.28 |
| chr5_140852570_140852762 | 0.14 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
22235 |
0.18 |
| chr13_114659678_114659829 | 0.14 |
4930544M13Rik |
RIKEN cDNA 4930544M13 gene |
52516 |
0.13 |
| chr18_54970945_54971399 | 0.14 |
Zfp608 |
zinc finger protein 608 |
14415 |
0.2 |
| chr11_23954708_23954885 | 0.13 |
Gm12062 |
predicted gene 12062 |
25942 |
0.16 |
| chr3_157748470_157748625 | 0.13 |
Gm33466 |
predicted gene, 33466 |
22139 |
0.2 |
| chr5_53046104_53046266 | 0.13 |
Gm17182 |
predicted gene 17182 |
3003 |
0.21 |
| chr19_21737626_21737804 | 0.13 |
Gm50129 |
predicted gene, 50129 |
8445 |
0.21 |
| chr6_104028580_104028747 | 0.13 |
Gm21054 |
predicted gene, 21054 |
64024 |
0.15 |
| chr5_117191658_117191809 | 0.13 |
n-R5s174 |
nuclear encoded rRNA 5S 174 |
4822 |
0.17 |
| chr6_128497203_128497354 | 0.13 |
Pzp |
PZP, alpha-2-macroglobulin like |
2055 |
0.17 |
| chr3_89280151_89280307 | 0.13 |
Efna1 |
ephrin A1 |
584 |
0.47 |
| chr9_85248224_85248375 | 0.13 |
Gm48831 |
predicted gene, 48831 |
8258 |
0.19 |
| chr8_85032460_85032611 | 0.13 |
Tnpo2 |
transportin 2 (importin 3, karyopherin beta 2b) |
4380 |
0.07 |
| chr6_116264926_116265157 | 0.13 |
Zfand4 |
zinc finger, AN1-type domain 4 |
776 |
0.4 |
| chr11_62487650_62487823 | 0.13 |
Gm12278 |
predicted gene 12278 |
4939 |
0.13 |
| chr12_108651085_108651241 | 0.13 |
Mir342 |
microRNA 342 |
7457 |
0.15 |
| chr5_63936466_63936784 | 0.13 |
Rell1 |
RELT-like 1 |
5711 |
0.18 |
| chr3_132863960_132864140 | 0.13 |
Gm29811 |
predicted gene, 29811 |
14546 |
0.14 |
| chr16_4029500_4029663 | 0.13 |
Dnase1 |
deoxyribonuclease I |
7361 |
0.1 |
| chr8_110941639_110941803 | 0.13 |
St3gal2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
817 |
0.47 |
| chr5_134639466_134639692 | 0.13 |
Eif4h |
eukaryotic translation initiation factor 4H |
89 |
0.95 |
| chr4_99102964_99103128 | 0.13 |
Gm12852 |
predicted gene 12852 |
732 |
0.62 |
| chr10_10472367_10472528 | 0.13 |
Adgb |
androglobin |
121 |
0.97 |
| chr16_30060579_30060767 | 0.13 |
Hes1 |
hes family bHLH transcription factor 1 |
3711 |
0.19 |
| chr1_93124444_93124635 | 0.13 |
Gm28086 |
predicted gene 28086 |
436 |
0.76 |
| chr5_107837254_107837906 | 0.13 |
Evi5 |
ecotropic viral integration site 5 |
100 |
0.94 |
| chr14_20345992_20346164 | 0.13 |
Ecd |
ecdysoneless cell cycle regulator |
1972 |
0.19 |
| chr9_51994808_51995091 | 0.13 |
Gm6981 |
predicted pseudogene 6981 |
8332 |
0.18 |
| chr1_4857561_4857736 | 0.13 |
Tcea1 |
transcription elongation factor A (SII) 1 |
166 |
0.93 |
| chr10_24948485_24948668 | 0.12 |
Gm36172 |
predicted gene, 36172 |
20957 |
0.13 |
| chr8_11486156_11486314 | 0.12 |
Rab20 |
RAB20, member RAS oncogene family |
7525 |
0.12 |
| chrX_129949506_129949684 | 0.12 |
Diaph2 |
diaphanous related formin 2 |
22938 |
0.28 |
| chr13_45637816_45638014 | 0.12 |
Gmpr |
guanosine monophosphate reductase |
91762 |
0.08 |
| chr1_86479051_86479408 | 0.12 |
Rpl30-ps6 |
ribosomal protein L30, pseudogene 6 |
5570 |
0.15 |
| chr7_44749310_44749461 | 0.12 |
Vrk3 |
vaccinia related kinase 3 |
442 |
0.55 |
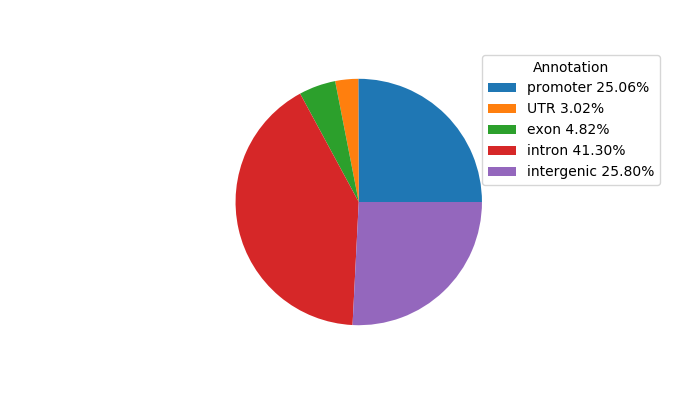
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.4 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0034823 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0018637 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |