Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
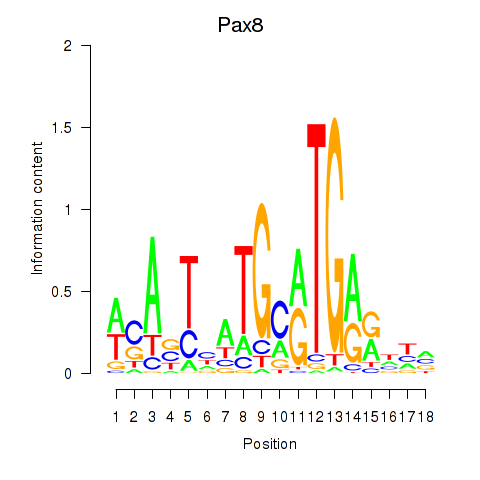
Results for Pax8
Z-value: 1.66
Transcription factors associated with Pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax8
|
ENSMUSG00000026976.9 | paired box 8 |
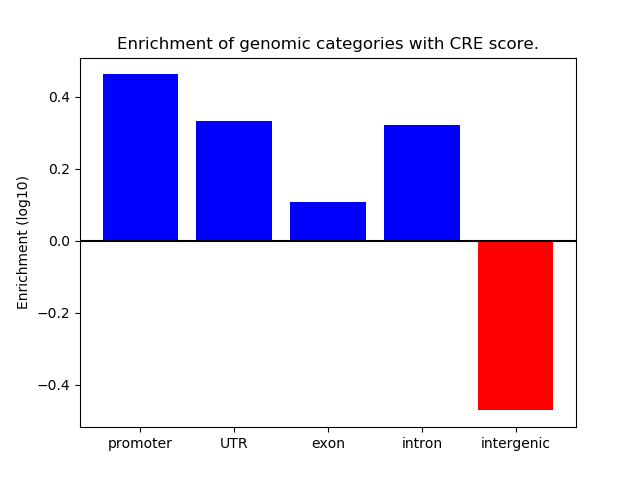
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_24474146_24474302 | Pax8 | 873 | 0.502829 | -0.73 | 9.9e-02 | Click! |
| chr2_24460542_24460763 | Pax8 | 14445 | 0.126804 | 0.58 | 2.3e-01 | Click! |
| chr2_24415026_24415177 | Pax8 | 7981 | 0.142348 | -0.47 | 3.5e-01 | Click! |
| chr2_24479227_24479538 | Pax8 | 3783 | 0.159818 | -0.31 | 5.4e-01 | Click! |
| chr2_24469183_24469359 | Pax8 | 5826 | 0.145448 | 0.18 | 7.3e-01 | Click! |
Activity of the Pax8 motif across conditions
Conditions sorted by the z-value of the Pax8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
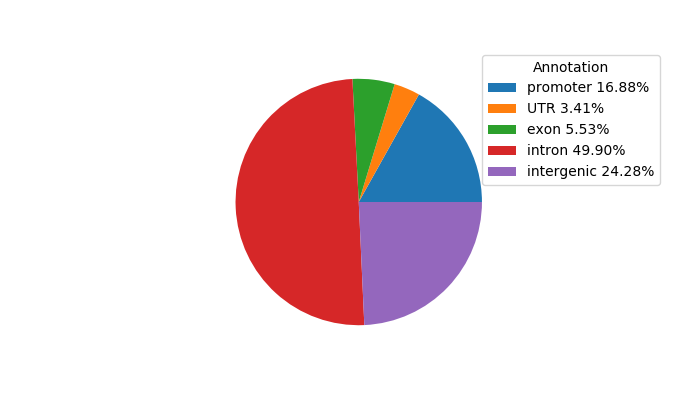
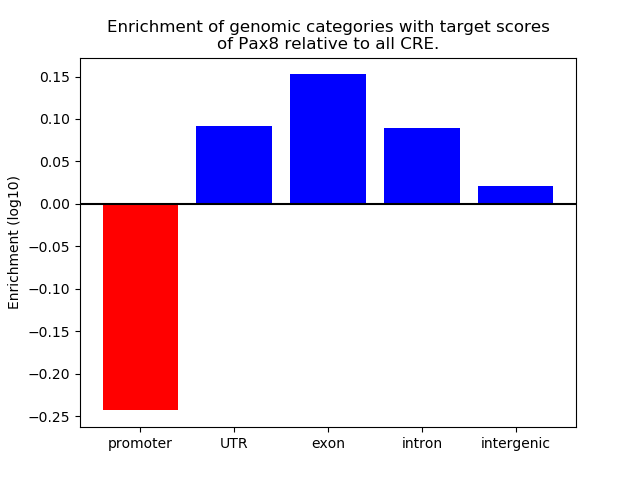
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_95144816_95145075 | 1.04 |
Gm29684 |
predicted gene, 29684 |
880 |
0.54 |
| chr10_111216225_111216427 | 0.93 |
Osbpl8 |
oxysterol binding protein-like 8 |
17303 |
0.16 |
| chr9_74886495_74886646 | 0.92 |
Onecut1 |
one cut domain, family member 1 |
20086 |
0.14 |
| chr11_16862611_16863177 | 0.90 |
Egfr |
epidermal growth factor receptor |
15256 |
0.19 |
| chr9_106232444_106232608 | 0.86 |
Alas1 |
aminolevulinic acid synthase 1 |
4558 |
0.11 |
| chr2_58773576_58773851 | 0.74 |
Upp2 |
uridine phosphorylase 2 |
8388 |
0.21 |
| chr6_136954641_136954792 | 0.73 |
Pde6h |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
183 |
0.93 |
| chr19_40165502_40165757 | 0.72 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
21657 |
0.14 |
| chr13_63665698_63665976 | 0.67 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr4_76942367_76942518 | 0.65 |
Gm11246 |
predicted gene 11246 |
15131 |
0.21 |
| chrX_85734642_85734808 | 0.63 |
Gk |
glycerol kinase |
2885 |
0.21 |
| chr19_40168031_40168182 | 0.61 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
19180 |
0.15 |
| chr8_76541013_76541167 | 0.60 |
Gm27355 |
predicted gene, 27355 |
78192 |
0.1 |
| chr11_16874000_16874630 | 0.60 |
Egfr |
epidermal growth factor receptor |
3835 |
0.25 |
| chr11_94239763_94239948 | 0.59 |
Wfikkn2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
2852 |
0.23 |
| chr5_114528485_114528658 | 0.57 |
Gm13790 |
predicted gene 13790 |
22718 |
0.14 |
| chr2_8124301_8124468 | 0.57 |
Gm13254 |
predicted gene 13254 |
23481 |
0.29 |
| chr10_80239863_80240052 | 0.53 |
Pwwp3a |
PWWP domain containing 3A, DNA repair factor |
1581 |
0.18 |
| chr12_103947018_103947616 | 0.53 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
9581 |
0.11 |
| chr3_63296372_63296537 | 0.52 |
Mme |
membrane metallo endopeptidase |
359 |
0.92 |
| chr8_84723608_84723805 | 0.51 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
699 |
0.52 |
| chr13_46154168_46154319 | 0.51 |
Gm10113 |
predicted gene 10113 |
36803 |
0.19 |
| chr19_44398143_44398357 | 0.50 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
8440 |
0.15 |
| chr4_104908553_104908715 | 0.50 |
Fyb2 |
FYN binding protein 2 |
4822 |
0.23 |
| chr1_50806813_50807031 | 0.50 |
Gm28321 |
predicted gene 28321 |
8453 |
0.28 |
| chr4_107999051_107999218 | 0.50 |
Slc1a7 |
solute carrier family 1 (glutamate transporter), member 7 |
10883 |
0.13 |
| chr1_23290878_23291029 | 0.49 |
Gm27028 |
predicted gene, 27028 |
584 |
0.46 |
| chr5_125482875_125483080 | 0.49 |
Gm27551 |
predicted gene, 27551 |
3600 |
0.16 |
| chr3_129450558_129450716 | 0.49 |
Rpl7a-ps7 |
ribosomal protein L7A, pseudogene 7 |
11335 |
0.17 |
| chr8_66494561_66494830 | 0.49 |
Gm32568 |
predicted gene, 32568 |
364 |
0.85 |
| chr8_61436826_61436991 | 0.49 |
Cbr4 |
carbonyl reductase 4 |
50826 |
0.12 |
| chr9_106231818_106232229 | 0.48 |
Alas1 |
aminolevulinic acid synthase 1 |
5061 |
0.11 |
| chr13_58962693_58962856 | 0.48 |
Ntrk2 |
neurotrophic tyrosine kinase, receptor, type 2 |
91039 |
0.06 |
| chrX_142506860_142507020 | 0.48 |
Gm25915 |
predicted gene, 25915 |
21493 |
0.16 |
| chr11_16894023_16894174 | 0.47 |
Egfr |
epidermal growth factor receptor |
11087 |
0.19 |
| chr1_21245041_21245248 | 0.47 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4515 |
0.13 |
| chr5_43310353_43310569 | 0.46 |
Gm43020 |
predicted gene 43020 |
44798 |
0.11 |
| chr5_148631104_148631264 | 0.46 |
Gm29815 |
predicted gene, 29815 |
17540 |
0.19 |
| chr14_120256809_120256970 | 0.45 |
Mbnl2 |
muscleblind like splicing factor 2 |
18780 |
0.23 |
| chr18_8864151_8864321 | 0.45 |
Gm37148 |
predicted gene, 37148 |
63999 |
0.13 |
| chr18_7657709_7657860 | 0.45 |
Mpp7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
30918 |
0.17 |
| chr16_11164937_11165106 | 0.45 |
Gm24145 |
predicted gene, 24145 |
2246 |
0.15 |
| chr2_134828255_134828422 | 0.45 |
Gm14036 |
predicted gene 14036 |
24389 |
0.2 |
| chr15_83452182_83452447 | 0.44 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
12238 |
0.15 |
| chr14_63172567_63172724 | 0.44 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
5148 |
0.16 |
| chr9_74894770_74895613 | 0.43 |
Onecut1 |
one cut domain, family member 1 |
28707 |
0.13 |
| chr15_3428785_3428957 | 0.43 |
Ghr |
growth hormone receptor |
42773 |
0.18 |
| chr12_104343910_104344587 | 0.43 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
5762 |
0.12 |
| chr1_87764160_87764454 | 0.42 |
Atg16l1 |
autophagy related 16-like 1 (S. cerevisiae) |
2594 |
0.19 |
| chr11_16862337_16862489 | 0.42 |
Egfr |
epidermal growth factor receptor |
15737 |
0.19 |
| chr1_21250942_21251436 | 0.42 |
Gsta3 |
glutathione S-transferase, alpha 3 |
2332 |
0.18 |
| chr14_33058246_33058397 | 0.42 |
Wdfy4 |
WD repeat and FYVE domain containing 4 |
34984 |
0.17 |
| chr7_107574186_107574390 | 0.42 |
Olfml1 |
olfactomedin-like 1 |
6522 |
0.16 |
| chr14_69626858_69627238 | 0.41 |
Gm37513 |
predicted gene, 37513 |
11002 |
0.11 |
| chr14_57106958_57107164 | 0.41 |
Gjb2 |
gap junction protein, beta 2 |
2359 |
0.23 |
| chr10_24258031_24258221 | 0.41 |
Moxd1 |
monooxygenase, DBH-like 1 |
34581 |
0.16 |
| chr4_62703366_62703517 | 0.41 |
Gm11210 |
predicted gene 11210 |
2540 |
0.23 |
| chr19_21626908_21627090 | 0.41 |
1110059E24Rik |
RIKEN cDNA 1110059E24 gene |
3769 |
0.26 |
| chr1_127899545_127899941 | 0.40 |
Rab3gap1 |
RAB3 GTPase activating protein subunit 1 |
1170 |
0.47 |
| chr6_119584715_119585189 | 0.40 |
Gm44317 |
predicted gene, 44317 |
14011 |
0.2 |
| chr14_69408593_69408964 | 0.40 |
Gm38378 |
predicted gene, 38378 |
11007 |
0.11 |
| chr12_83935202_83935387 | 0.40 |
Numb |
NUMB endocytic adaptor protein |
13360 |
0.1 |
| chr13_107469451_107469885 | 0.39 |
AI197445 |
expressed sequence AI197445 |
154 |
0.97 |
| chr16_24878031_24878240 | 0.38 |
Gm22672 |
predicted gene, 22672 |
6038 |
0.25 |
| chr19_44394314_44394509 | 0.38 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12279 |
0.14 |
| chr5_125528530_125528687 | 0.38 |
Tmem132b |
transmembrane protein 132B |
3166 |
0.22 |
| chr18_55182860_55183019 | 0.38 |
Gm22597 |
predicted gene, 22597 |
3099 |
0.29 |
| chr16_43168807_43168974 | 0.38 |
Gm15712 |
predicted gene 15712 |
15683 |
0.21 |
| chr14_7842334_7842485 | 0.38 |
Flnb |
filamin, beta |
24452 |
0.15 |
| chr19_40160539_40160690 | 0.38 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
26672 |
0.14 |
| chr2_58773911_58774203 | 0.37 |
Upp2 |
uridine phosphorylase 2 |
8732 |
0.21 |
| chr8_41202065_41202231 | 0.37 |
Fgl1 |
fibrinogen-like protein 1 |
13005 |
0.17 |
| chr17_57205198_57205712 | 0.37 |
Tnfsf14 |
tumor necrosis factor (ligand) superfamily, member 14 |
11278 |
0.1 |
| chr2_52579266_52579417 | 0.37 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
20774 |
0.18 |
| chr11_51753632_51753825 | 0.37 |
Sec24a |
Sec24 related gene family, member A (S. cerevisiae) |
2483 |
0.22 |
| chr11_16588986_16589143 | 0.36 |
Gm12663 |
predicted gene 12663 |
47002 |
0.12 |
| chr8_36290613_36290918 | 0.36 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
41249 |
0.14 |
| chr12_57537784_57538085 | 0.36 |
Foxa1 |
forkhead box A1 |
8187 |
0.15 |
| chr5_99474871_99475183 | 0.36 |
Gm35172 |
predicted gene, 35172 |
33362 |
0.17 |
| chr14_30542112_30542288 | 0.36 |
Tkt |
transketolase |
6159 |
0.16 |
| chr2_31487515_31488471 | 0.36 |
Ass1 |
argininosuccinate synthetase 1 |
9779 |
0.18 |
| chr8_114152687_114152971 | 0.35 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
19187 |
0.25 |
| chr2_31497033_31497351 | 0.35 |
Ass1 |
argininosuccinate synthetase 1 |
580 |
0.75 |
| chr12_104083020_104083260 | 0.35 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr1_67165771_67165961 | 0.35 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
42840 |
0.16 |
| chr19_44393747_44393961 | 0.35 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12836 |
0.14 |
| chr11_16814206_16814660 | 0.35 |
Egfros |
epidermal growth factor receptor, opposite strand |
16269 |
0.21 |
| chr10_66914558_66914709 | 0.35 |
1110002J07Rik |
RIKEN cDNA 1110002J07 gene |
3106 |
0.21 |
| chr1_77279465_77279665 | 0.35 |
Epha4 |
Eph receptor A4 |
98042 |
0.08 |
| chr2_58791427_58791720 | 0.35 |
Upp2 |
uridine phosphorylase 2 |
26248 |
0.18 |
| chr3_138226845_138227007 | 0.35 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
1125 |
0.38 |
| chr15_10200971_10201518 | 0.34 |
Prlr |
prolactin receptor |
12221 |
0.28 |
| chr5_92097101_92097294 | 0.34 |
G3bp2 |
GTPase activating protein (SH3 domain) binding protein 2 |
13478 |
0.12 |
| chr17_8810128_8810417 | 0.34 |
Pde10a |
phosphodiesterase 10A |
7244 |
0.26 |
| chr11_90685296_90685460 | 0.34 |
Tom1l1 |
target of myb1-like 1 (chicken) |
2201 |
0.34 |
| chr12_80010993_80011167 | 0.33 |
Gm8275 |
predicted gene 8275 |
31229 |
0.14 |
| chr6_71812965_71813119 | 0.33 |
Gm44770 |
predicted gene 44770 |
10154 |
0.1 |
| chr19_40174636_40174787 | 0.33 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
12575 |
0.15 |
| chr9_21689288_21689439 | 0.33 |
Gm26511 |
predicted gene, 26511 |
34087 |
0.09 |
| chr6_50091657_50091996 | 0.33 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
18415 |
0.22 |
| chr17_46051138_46051309 | 0.33 |
Vegfa |
vascular endothelial growth factor A |
18854 |
0.13 |
| chr16_65666527_65666684 | 0.32 |
Gm49633 |
predicted gene, 49633 |
53186 |
0.14 |
| chr19_55394273_55394505 | 0.32 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
13375 |
0.25 |
| chr1_74970898_74971209 | 0.32 |
Gm37744 |
predicted gene, 37744 |
16801 |
0.11 |
| chr4_84540692_84540849 | 0.32 |
Bnc2 |
basonuclin 2 |
5520 |
0.32 |
| chr2_20909493_20909665 | 0.32 |
Arhgap21 |
Rho GTPase activating protein 21 |
5115 |
0.23 |
| chr3_18210165_18210316 | 0.32 |
Gm23686 |
predicted gene, 23686 |
32615 |
0.17 |
| chr1_67111462_67111613 | 0.31 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
11489 |
0.23 |
| chr2_52578719_52578880 | 0.31 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
20232 |
0.18 |
| chr2_92424112_92424429 | 0.31 |
Cry2 |
cryptochrome 2 (photolyase-like) |
8122 |
0.11 |
| chr12_75566070_75566284 | 0.31 |
Gm47689 |
predicted gene, 47689 |
13688 |
0.18 |
| chr2_160361611_160362000 | 0.31 |
Mafb |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
5260 |
0.27 |
| chr13_43530948_43531160 | 0.31 |
Gm32939 |
predicted gene, 32939 |
834 |
0.53 |
| chr11_7815566_7815774 | 0.31 |
Gm27393 |
predicted gene, 27393 |
70835 |
0.13 |
| chr19_43496566_43496970 | 0.31 |
Cnnm1 |
cyclin M1 |
3252 |
0.16 |
| chr5_112242342_112242634 | 0.31 |
Cryba4 |
crystallin, beta A4 |
8674 |
0.11 |
| chr8_64843591_64843742 | 0.30 |
Klhl2 |
kelch-like 2, Mayven |
1469 |
0.34 |
| chr2_160763946_160764097 | 0.30 |
Plcg1 |
phospholipase C, gamma 1 |
3903 |
0.2 |
| chr1_36095531_36095713 | 0.30 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
6068 |
0.15 |
| chr4_104763940_104764114 | 0.30 |
C8b |
complement component 8, beta polypeptide |
2290 |
0.4 |
| chr9_74294555_74294732 | 0.30 |
Wdr72 |
WD repeat domain 72 |
20509 |
0.23 |
| chrX_166678508_166678678 | 0.30 |
Gm1720 |
predicted gene 1720 |
16568 |
0.21 |
| chr7_97413837_97414007 | 0.30 |
Thrsp |
thyroid hormone responsive |
3597 |
0.16 |
| chr3_94708222_94708411 | 0.30 |
Selenbp2 |
selenium binding protein 2 |
14657 |
0.1 |
| chr8_72665230_72665413 | 0.30 |
Nwd1 |
NACHT and WD repeat domain containing 1 |
8235 |
0.15 |
| chr10_111150128_111150279 | 0.30 |
Gm8873 |
predicted gene 8873 |
6013 |
0.14 |
| chr7_75457902_75458053 | 0.30 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
2109 |
0.28 |
| chr15_58983651_58983950 | 0.29 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
336 |
0.87 |
| chr19_26778139_26778521 | 0.29 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
6734 |
0.21 |
| chr4_138516557_138516728 | 0.29 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
62328 |
0.08 |
| chr11_16828832_16828991 | 0.29 |
Egfros |
epidermal growth factor receptor, opposite strand |
1791 |
0.4 |
| chr11_99105082_99105257 | 0.29 |
Tns4 |
tensin 4 |
15863 |
0.14 |
| chr13_40888163_40888333 | 0.29 |
Gcnt2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
1055 |
0.36 |
| chr6_66996154_66996403 | 0.28 |
Gm36816 |
predicted gene, 36816 |
12135 |
0.12 |
| chr3_28703876_28704177 | 0.28 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
6009 |
0.18 |
| chr17_24882806_24883274 | 0.28 |
Nubp2 |
nucleotide binding protein 2 |
2577 |
0.11 |
| chr1_61971987_61972299 | 0.28 |
Gm29640 |
predicted gene 29640 |
9064 |
0.27 |
| chr8_105091387_105091623 | 0.28 |
Ces3b |
carboxylesterase 3B |
2886 |
0.14 |
| chr5_90471470_90471733 | 0.28 |
Alb |
albumin |
8901 |
0.15 |
| chr3_19978796_19978963 | 0.27 |
Cp |
ceruloplasmin |
1749 |
0.34 |
| chr11_11844304_11844501 | 0.27 |
Ddc |
dopa decarboxylase |
8122 |
0.18 |
| chr2_72222531_72222771 | 0.27 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
9651 |
0.17 |
| chr1_21262098_21262745 | 0.27 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8900 |
0.11 |
| chr6_27967952_27968118 | 0.27 |
Mir592 |
microRNA 592 |
31285 |
0.23 |
| chr6_66963164_66963414 | 0.27 |
Gm36816 |
predicted gene, 36816 |
45124 |
0.08 |
| chr19_58738230_58738402 | 0.27 |
Pnliprp1 |
pancreatic lipase related protein 1 |
1326 |
0.36 |
| chr16_42914859_42915016 | 0.27 |
Gm19522 |
predicted gene, 19522 |
2872 |
0.25 |
| chr3_18156058_18156209 | 0.27 |
Gm23686 |
predicted gene, 23686 |
21492 |
0.21 |
| chr2_122260835_122261566 | 0.27 |
Duox2 |
dual oxidase 2 |
24369 |
0.09 |
| chr10_86691292_86691645 | 0.27 |
Gm15344 |
predicted gene 15344 |
1627 |
0.16 |
| chr11_16798476_16798627 | 0.27 |
Egfros |
epidermal growth factor receptor, opposite strand |
32151 |
0.16 |
| chr8_9027955_9028106 | 0.27 |
Gm44515 |
predicted gene 44515 |
95331 |
0.06 |
| chr17_84734946_84735097 | 0.27 |
Lrpprc |
leucine-rich PPR-motif containing |
3837 |
0.21 |
| chr18_9842900_9843072 | 0.26 |
Gm23637 |
predicted gene, 23637 |
15867 |
0.13 |
| chr6_149169218_149169449 | 0.26 |
Amn1 |
antagonist of mitotic exit network 1 |
4038 |
0.17 |
| chr9_80256438_80256780 | 0.26 |
Myo6 |
myosin VI |
5438 |
0.23 |
| chr11_16763486_16763658 | 0.26 |
Egfr |
epidermal growth factor receptor |
11342 |
0.2 |
| chr3_95322811_95323271 | 0.26 |
Cers2 |
ceramide synthase 2 |
2101 |
0.14 |
| chr6_115238722_115238899 | 0.26 |
Syn2 |
synapsin II |
184 |
0.94 |
| chr10_20749070_20749231 | 0.26 |
Gm48651 |
predicted gene, 48651 |
6967 |
0.23 |
| chr13_69532134_69532307 | 0.26 |
Tent4a |
terminal nucleotidyltransferase 4A |
212 |
0.89 |
| chr8_64777953_64778119 | 0.26 |
Klhl2 |
kelch-like 2, Mayven |
21628 |
0.14 |
| chr15_77384660_77384984 | 0.26 |
Gm20688 |
predicted gene 20688 |
7559 |
0.1 |
| chr1_163009105_163009274 | 0.26 |
Fmo3 |
flavin containing monooxygenase 3 |
24661 |
0.14 |
| chr19_5794205_5794418 | 0.26 |
Malat1 |
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
2641 |
0.11 |
| chr14_75284686_75284868 | 0.26 |
Zc3h13 |
zinc finger CCCH type containing 13 |
391 |
0.82 |
| chr17_84226805_84226999 | 0.26 |
Thada |
thyroid adenoma associated |
2958 |
0.24 |
| chr12_100421631_100422293 | 0.26 |
Gm48381 |
predicted gene, 48381 |
22698 |
0.15 |
| chr15_3468356_3468528 | 0.26 |
Ghr |
growth hormone receptor |
3202 |
0.36 |
| chr9_42486993_42487198 | 0.25 |
Gm16322 |
predicted gene 16322 |
11521 |
0.18 |
| chr11_21406163_21406488 | 0.25 |
Ugp2 |
UDP-glucose pyrophosphorylase 2 |
35124 |
0.11 |
| chr19_40183641_40183792 | 0.25 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
3570 |
0.19 |
| chr3_60965282_60965682 | 0.25 |
P2ry1 |
purinergic receptor P2Y, G-protein coupled 1 |
37313 |
0.16 |
| chr18_4930037_4930197 | 0.25 |
Svil |
supervillin |
8391 |
0.3 |
| chr3_130440388_130440664 | 0.25 |
Col25a1 |
collagen, type XXV, alpha 1 |
38290 |
0.17 |
| chr19_38028254_38028422 | 0.25 |
Myof |
myoferlin |
15013 |
0.15 |
| chr10_99267793_99268059 | 0.25 |
Gm48089 |
predicted gene, 48089 |
30 |
0.95 |
| chr9_21837693_21838006 | 0.25 |
Angptl8 |
angiopoietin-like 8 |
2339 |
0.17 |
| chr14_120338170_120338321 | 0.25 |
Mbnl2 |
muscleblind like splicing factor 2 |
31789 |
0.19 |
| chr5_116390234_116390471 | 0.25 |
4930562A09Rik |
RIKEN cDNA 4930562A09 gene |
2448 |
0.2 |
| chr9_96893333_96893484 | 0.25 |
Pxylp1 |
2-phosphoxylose phosphatase 1 |
739 |
0.57 |
| chr1_67143038_67143247 | 0.25 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
20116 |
0.22 |
| chr1_67214555_67214706 | 0.25 |
Gm15668 |
predicted gene 15668 |
34570 |
0.17 |
| chr4_45411492_45411708 | 0.25 |
Slc25a51 |
solute carrier family 25, member 51 |
2834 |
0.21 |
| chr2_31503965_31504231 | 0.25 |
Ass1 |
argininosuccinate synthetase 1 |
3372 |
0.23 |
| chr5_53992838_53993011 | 0.25 |
Stim2 |
stromal interaction molecule 2 |
5575 |
0.27 |
| chr11_16795963_16796156 | 0.25 |
Egfros |
epidermal growth factor receptor, opposite strand |
34643 |
0.15 |
| chr2_177901070_177901508 | 0.25 |
Gm14327 |
predicted gene 14327 |
2996 |
0.19 |
| chr2_134503152_134503303 | 0.25 |
Hao1 |
hydroxyacid oxidase 1, liver |
51080 |
0.18 |
| chr14_17720570_17720721 | 0.24 |
Gm48320 |
predicted gene, 48320 |
50477 |
0.16 |
| chr8_93183400_93183567 | 0.24 |
Gm45909 |
predicted gene 45909 |
7875 |
0.13 |
| chr11_16818941_16819203 | 0.24 |
Egfros |
epidermal growth factor receptor, opposite strand |
11630 |
0.22 |
| chr9_55322522_55322738 | 0.24 |
Nrg4 |
neuregulin 4 |
2557 |
0.25 |
| chr13_4230959_4231234 | 0.24 |
Akr1c19 |
aldo-keto reductase family 1, member C19 |
2396 |
0.22 |
| chr8_93183969_93184163 | 0.24 |
Gm45909 |
predicted gene 45909 |
7292 |
0.14 |
| chr7_101106567_101106718 | 0.24 |
Fchsd2 |
FCH and double SH3 domains 2 |
2091 |
0.29 |
| chr9_106888296_106888447 | 0.24 |
Rbm15b |
RNA binding motif protein 15B |
943 |
0.37 |
| chr3_121852936_121853213 | 0.24 |
Gm42593 |
predicted gene 42593 |
9768 |
0.15 |
| chr11_120809505_120809952 | 0.24 |
Fasn |
fatty acid synthase |
27 |
0.95 |
| chr11_53278374_53278525 | 0.24 |
Hspa4 |
heat shock protein 4 |
11208 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.8 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.2 | 0.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 1.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.2 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.4 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0070836 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.1 | GO:0003176 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of RNA catabolic process(GO:1902369) negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.0 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.0 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0080030 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0034929 | fluorene oxygenase activity(GO:0018585) mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME DNA REPLICATION | Genes involved in DNA Replication |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.0 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |