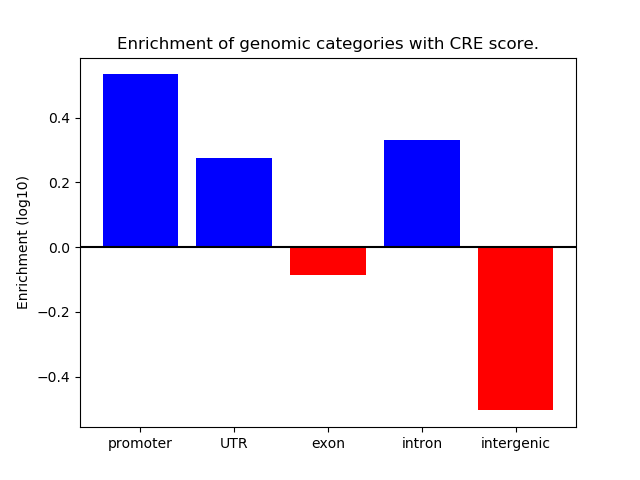
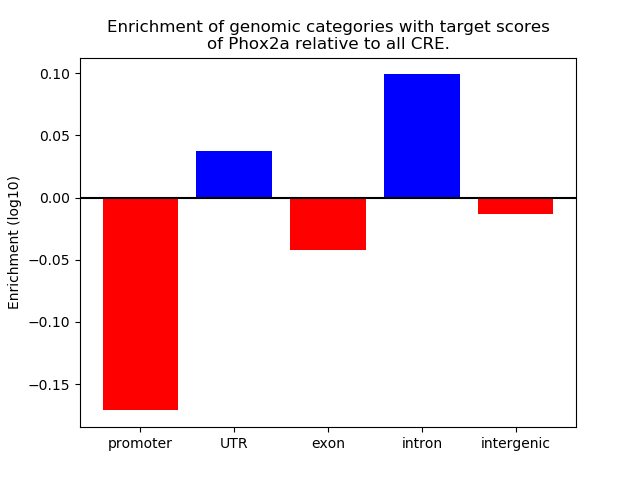
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
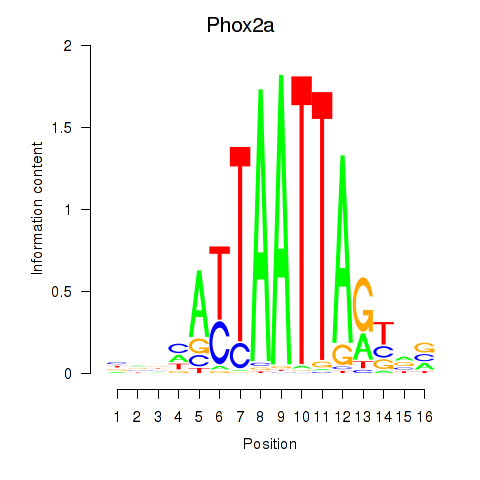
Results for Phox2a
Z-value: 1.02
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSMUSG00000007946.9 | paired-like homeobox 2a |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_101822093_101822270 | Phox2a | 1435 | 0.218398 | -0.75 | 8.3e-02 | Click! |
| chr7_101821639_101821825 | Phox2a | 986 | 0.333450 | -0.50 | 3.1e-01 | Click! |
Activity of the Phox2a motif across conditions
Conditions sorted by the z-value of the Phox2a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_18048778_18049088 | 1.25 |
Skida1 |
SKI/DACH domain containing 1 |
98 |
0.94 |
| chr17_80022007_80022158 | 1.03 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr19_44401346_44401497 | 0.93 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5269 |
0.16 |
| chr12_75531904_75532183 | 0.92 |
Ppp2r5e |
protein phosphatase 2, regulatory subunit B', epsilon |
17631 |
0.18 |
| chr1_51740463_51740849 | 0.86 |
Gm28055 |
predicted gene 28055 |
6287 |
0.22 |
| chr11_30792834_30792985 | 0.83 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
18628 |
0.14 |
| chr7_65902304_65902456 | 0.83 |
Pcsk6 |
proprotein convertase subtilisin/kexin type 6 |
39948 |
0.14 |
| chr6_52610697_52610915 | 0.82 |
Gm44434 |
predicted gene, 44434 |
7561 |
0.15 |
| chr4_44777019_44777170 | 0.77 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
16650 |
0.16 |
| chr7_84113279_84113524 | 0.77 |
Cemip |
cell migration inducing protein, hyaluronan binding |
26899 |
0.14 |
| chr19_3864646_3865022 | 0.76 |
Gm50383 |
predicted gene, 50383 |
2839 |
0.13 |
| chr19_3872249_3872405 | 0.73 |
Chka |
choline kinase alpha |
2337 |
0.14 |
| chr7_30967090_30967447 | 0.65 |
Lsr |
lipolysis stimulated lipoprotein receptor |
6054 |
0.07 |
| chr13_117745874_117746186 | 0.65 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
26019 |
0.26 |
| chr2_158093657_158093889 | 0.64 |
2010009K17Rik |
RIKEN cDNA 2010009K17 gene |
1976 |
0.28 |
| chr1_23237656_23237879 | 0.63 |
Gm29506 |
predicted gene 29506 |
2094 |
0.23 |
| chr18_39438249_39438499 | 0.60 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
48858 |
0.13 |
| chr8_12804734_12804948 | 0.60 |
Atp11a |
ATPase, class VI, type 11A |
6575 |
0.17 |
| chr12_28636385_28636550 | 0.58 |
Rps7 |
ribosomal protein S7 |
514 |
0.71 |
| chr12_51274477_51274668 | 0.56 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
22915 |
0.22 |
| chr12_73861884_73862127 | 0.56 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr4_106354190_106354378 | 0.56 |
Usp24 |
ubiquitin specific peptidase 24 |
38050 |
0.13 |
| chr4_15212980_15213172 | 0.55 |
Tmem64 |
transmembrane protein 64 |
52755 |
0.13 |
| chr10_4618620_4618773 | 0.55 |
Esr1 |
estrogen receptor 1 (alpha) |
6675 |
0.25 |
| chr2_67977965_67978321 | 0.54 |
Gm37964 |
predicted gene, 37964 |
79547 |
0.1 |
| chr12_73341772_73342027 | 0.54 |
Slc38a6 |
solute carrier family 38, member 6 |
2468 |
0.25 |
| chr11_22844106_22844418 | 0.53 |
Gm23772 |
predicted gene, 23772 |
2606 |
0.17 |
| chr1_13368915_13369352 | 0.52 |
Ncoa2 |
nuclear receptor coactivator 2 |
3296 |
0.16 |
| chr19_48883437_48883771 | 0.51 |
Gm50436 |
predicted gene, 50436 |
72468 |
0.11 |
| chr9_112785054_112785229 | 0.51 |
Gm24957 |
predicted gene, 24957 |
262612 |
0.02 |
| chr6_119380906_119381057 | 0.50 |
Adipor2 |
adiponectin receptor 2 |
7705 |
0.21 |
| chr17_88605968_88606119 | 0.49 |
Gm9406 |
predicted gene 9406 |
2501 |
0.24 |
| chr9_112123334_112123526 | 0.49 |
Mir128-2 |
microRNA 128-2 |
4719 |
0.29 |
| chr16_92631807_92631997 | 0.48 |
Gm26626 |
predicted gene, 26626 |
19088 |
0.2 |
| chr13_118829008_118829159 | 0.48 |
Gm47335 |
predicted gene, 47335 |
29331 |
0.22 |
| chr8_35495669_35495888 | 0.47 |
Eri1 |
exoribonuclease 1 |
245 |
0.93 |
| chr12_85291673_85291832 | 0.47 |
Zc2hc1c |
zinc finger, C2HC-type containing 1C |
3161 |
0.14 |
| chr5_138080510_138080792 | 0.47 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr19_44400352_44400581 | 0.47 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6224 |
0.15 |
| chr9_69333993_69334216 | 0.47 |
Gm15511 |
predicted gene 15511 |
30143 |
0.16 |
| chr6_122148793_122148971 | 0.46 |
Gm10319 |
predicted pseudogene 10319 |
12150 |
0.17 |
| chr17_17234200_17234351 | 0.46 |
Gm5165 |
predicted gene 5165 |
597 |
0.78 |
| chr9_53291516_53291691 | 0.45 |
Exph5 |
exophilin 5 |
10067 |
0.19 |
| chr18_3465417_3465568 | 0.45 |
Gm50088 |
predicted gene, 50088 |
1010 |
0.43 |
| chr3_131485378_131485553 | 0.45 |
Sgms2 |
sphingomyelin synthase 2 |
5014 |
0.28 |
| chr4_123990670_123991070 | 0.44 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr2_107628721_107628872 | 0.44 |
Gm9864 |
predicted gene 9864 |
28257 |
0.26 |
| chr15_82404193_82404671 | 0.44 |
Cyp2d10 |
cytochrome P450, family 2, subfamily d, polypeptide 10 |
1535 |
0.14 |
| chr16_65665507_65665824 | 0.43 |
Gm49633 |
predicted gene, 49633 |
52246 |
0.14 |
| chr5_87030613_87030972 | 0.43 |
Gm18635 |
predicted gene, 18635 |
16632 |
0.1 |
| chr17_84997546_84997697 | 0.42 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
3941 |
0.22 |
| chr2_150913728_150914090 | 0.42 |
Gins1 |
GINS complex subunit 1 (Psf1 homolog) |
1090 |
0.37 |
| chr13_93710959_93711124 | 0.42 |
Dmgdh |
dimethylglycine dehydrogenase precursor |
498 |
0.72 |
| chr19_7182336_7182545 | 0.41 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
18024 |
0.12 |
| chr5_134177966_134178117 | 0.41 |
Rcc1l |
reculator of chromosome condensation 1 like |
1267 |
0.3 |
| chr17_18321520_18321874 | 0.41 |
Vmn2r93 |
vomeronasal 2, receptor 93 |
23598 |
0.19 |
| chr6_38794603_38794986 | 0.41 |
Hipk2 |
homeodomain interacting protein kinase 2 |
23552 |
0.21 |
| chr10_31551438_31551618 | 0.41 |
Gm47693 |
predicted gene, 47693 |
5912 |
0.19 |
| chr11_42198938_42199110 | 0.41 |
Gabra1 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
16094 |
0.28 |
| chr14_65740190_65740357 | 0.41 |
Scara5 |
scavenger receptor class A, member 5 |
63796 |
0.1 |
| chr19_30093178_30093359 | 0.41 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
1307 |
0.48 |
| chr18_16822318_16822486 | 0.41 |
Gm15328 |
predicted gene 15328 |
5995 |
0.21 |
| chr6_6213107_6213286 | 0.41 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
3922 |
0.28 |
| chr19_6919936_6920100 | 0.41 |
Esrra |
estrogen related receptor, alpha |
1033 |
0.26 |
| chr2_125783158_125783336 | 0.40 |
Secisbp2l |
SECIS binding protein 2-like |
377 |
0.89 |
| chr17_81386211_81386362 | 0.40 |
Gm50044 |
predicted gene, 50044 |
15453 |
0.24 |
| chr11_4095549_4095867 | 0.40 |
Mtfp1 |
mitochondrial fission process 1 |
263 |
0.83 |
| chr4_109104769_109104920 | 0.39 |
Osbpl9 |
oxysterol binding protein-like 9 |
3187 |
0.27 |
| chr12_4220651_4220920 | 0.39 |
Gm48210 |
predicted gene, 48210 |
1317 |
0.27 |
| chr2_103461502_103462486 | 0.39 |
Elf5 |
E74-like factor 5 |
13256 |
0.17 |
| chr13_37756552_37756712 | 0.39 |
Gm31600 |
predicted gene, 31600 |
5116 |
0.16 |
| chr14_68299957_68300145 | 0.38 |
Gm47212 |
predicted gene, 47212 |
81798 |
0.09 |
| chr16_94606252_94606403 | 0.38 |
Gm15971 |
predicted gene 15971 |
14050 |
0.19 |
| chr16_78470224_78470428 | 0.38 |
Gm49603 |
predicted gene, 49603 |
10213 |
0.17 |
| chr17_33667328_33667604 | 0.38 |
Hnrnpm |
heterogeneous nuclear ribonucleoprotein M |
896 |
0.43 |
| chr12_111819707_111819858 | 0.38 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
3526 |
0.14 |
| chr2_160865219_160865594 | 0.38 |
Zhx3 |
zinc fingers and homeoboxes 3 |
5775 |
0.14 |
| chr10_20723719_20723889 | 0.38 |
Pde7b |
phosphodiesterase 7B |
892 |
0.65 |
| chr7_101838012_101838169 | 0.38 |
Inppl1 |
inositol polyphosphate phosphatase-like 1 |
37 |
0.7 |
| chr19_53904567_53904727 | 0.37 |
Pdcd4 |
programmed cell death 4 |
1231 |
0.39 |
| chr5_125526040_125526191 | 0.37 |
Tmem132b |
transmembrane protein 132B |
5659 |
0.17 |
| chr15_6218493_6218848 | 0.37 |
Gm23139 |
predicted gene, 23139 |
67322 |
0.11 |
| chr1_151154930_151155204 | 0.37 |
C730036E19Rik |
RIKEN cDNA C730036E19 gene |
17033 |
0.11 |
| chr3_94339066_94339223 | 0.36 |
Them5 |
thioesterase superfamily member 5 |
2955 |
0.11 |
| chr11_115919240_115919538 | 0.36 |
Recql5os1 |
RecQ-like 5, opposite strand 1 |
5638 |
0.09 |
| chr19_21654390_21654886 | 0.36 |
Abhd17b |
abhydrolase domain containing 17B |
1113 |
0.45 |
| chr16_13359323_13359477 | 0.36 |
Mrtfb |
myocardin related transcription factor B |
979 |
0.59 |
| chr13_37854787_37854958 | 0.36 |
Rreb1 |
ras responsive element binding protein 1 |
3062 |
0.27 |
| chr8_41069792_41069999 | 0.35 |
Mtus1 |
mitochondrial tumor suppressor 1 |
12881 |
0.16 |
| chr14_25916848_25916999 | 0.35 |
Tmem254a |
transmembrane protein 254a |
8865 |
0.12 |
| chr6_72120521_72121047 | 0.35 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr9_48787462_48787613 | 0.35 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
48408 |
0.15 |
| chr7_49458254_49458469 | 0.34 |
Nav2 |
neuron navigator 2 |
7601 |
0.23 |
| chr12_12940858_12941058 | 0.34 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
342 |
0.83 |
| chr13_23299091_23299367 | 0.34 |
Gm11333 |
predicted gene 11333 |
1180 |
0.26 |
| chr4_126764112_126764263 | 0.34 |
AU040320 |
expressed sequence AU040320 |
10361 |
0.13 |
| chr12_80336463_80336614 | 0.34 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
40154 |
0.1 |
| chr14_26056611_26056762 | 0.34 |
Tmem254c |
transmembrane protein 254c |
8855 |
0.11 |
| chr3_69543570_69543742 | 0.34 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
1224 |
0.48 |
| chr4_116838867_116839391 | 0.34 |
Rpl36-ps8 |
ribosomal protein L36, pseudogene 8 |
1092 |
0.3 |
| chr1_74003708_74004000 | 0.34 |
Tns1 |
tensin 1 |
2287 |
0.36 |
| chr18_39427173_39427361 | 0.34 |
Gm15337 |
predicted gene 15337 |
37842 |
0.16 |
| chr16_37566906_37567176 | 0.33 |
Rabl3 |
RAB, member RAS oncogene family-like 3 |
3307 |
0.18 |
| chr8_117738226_117738377 | 0.33 |
Gm31774 |
predicted gene, 31774 |
9959 |
0.14 |
| chr18_35306593_35306763 | 0.33 |
Sil1 |
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
6863 |
0.2 |
| chrX_38400123_38400274 | 0.33 |
Gm4853 |
predicted pseudogene 4853 |
3263 |
0.22 |
| chr9_119401641_119401816 | 0.33 |
Acvr2b |
activin receptor IIB |
390 |
0.77 |
| chr2_35098773_35098970 | 0.33 |
AI182371 |
expressed sequence AI182371 |
1813 |
0.3 |
| chr5_30286285_30286785 | 0.32 |
Drc1 |
dynein regulatory complex subunit 1 |
5147 |
0.17 |
| chr3_52854941_52855092 | 0.32 |
Gm30292 |
predicted gene, 30292 |
57231 |
0.11 |
| chr13_4205379_4205553 | 0.32 |
Akr1c13 |
aldo-keto reductase family 1, member C13 |
11608 |
0.13 |
| chr9_89704333_89704504 | 0.32 |
Tmed3 |
transmembrane p24 trafficking protein 3 |
405 |
0.82 |
| chr17_64607771_64607970 | 0.32 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr9_67840552_67841087 | 0.32 |
Vps13c |
vacuolar protein sorting 13C |
407 |
0.85 |
| chr6_144895356_144895507 | 0.32 |
Gm22792 |
predicted gene, 22792 |
91799 |
0.07 |
| chr5_147545326_147545477 | 0.32 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
6124 |
0.21 |
| chr9_62023104_62023499 | 0.32 |
Paqr5 |
progestin and adipoQ receptor family member V |
3500 |
0.27 |
| chr6_120318643_120318978 | 0.31 |
B4galnt3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
24251 |
0.15 |
| chr11_35750589_35750898 | 0.31 |
Pank3 |
pantothenate kinase 3 |
18741 |
0.16 |
| chr10_82740996_82741147 | 0.31 |
Nfyb |
nuclear transcription factor-Y beta |
10588 |
0.13 |
| chr6_33823830_33824315 | 0.31 |
Exoc4 |
exocyst complex component 4 |
38028 |
0.2 |
| chr12_75735898_75736072 | 0.31 |
Sgpp1 |
sphingosine-1-phosphate phosphatase 1 |
256 |
0.94 |
| chr5_64337855_64338006 | 0.31 |
Gm6044 |
predicted gene 6044 |
17674 |
0.13 |
| chr3_146778044_146778430 | 0.31 |
Prkacb |
protein kinase, cAMP dependent, catalytic, beta |
3179 |
0.23 |
| chr2_122737415_122737579 | 0.31 |
Bloc1s6os |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin, opposite strand |
865 |
0.39 |
| chr17_88614606_88614762 | 0.30 |
Gm9406 |
predicted gene 9406 |
6140 |
0.17 |
| chr4_152741969_152742142 | 0.30 |
Gm833 |
predicted gene 833 |
44515 |
0.17 |
| chr4_105198345_105198538 | 0.30 |
Plpp3 |
phospholipid phosphatase 3 |
41094 |
0.19 |
| chr6_65569857_65570008 | 0.30 |
Tnip3 |
TNFAIP3 interacting protein 3 |
20466 |
0.22 |
| chr6_37510078_37510463 | 0.29 |
Akr1d1 |
aldo-keto reductase family 1, member D1 |
19903 |
0.21 |
| chr3_19997345_19997496 | 0.29 |
Cp |
ceruloplasmin |
8268 |
0.18 |
| chr9_89882776_89882950 | 0.29 |
Rasgrf1 |
RAS protein-specific guanine nucleotide-releasing factor 1 |
27045 |
0.16 |
| chr15_16844404_16844562 | 0.29 |
Gm49127 |
predicted gene, 49127 |
6481 |
0.3 |
| chr14_119831250_119831463 | 0.29 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
32378 |
0.19 |
| chr5_124025292_124025465 | 0.29 |
Vps37b |
vacuolar protein sorting 37B |
6880 |
0.1 |
| chr11_120695384_120695739 | 0.29 |
Aspscr1 |
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
6251 |
0.07 |
| chr16_18071409_18071560 | 0.29 |
Dgcr6 |
DiGeorge syndrome critical region gene 6 |
2004 |
0.23 |
| chr14_20956319_20956501 | 0.29 |
Vcl |
vinculin |
27012 |
0.18 |
| chr12_77681559_77681753 | 0.28 |
4930458K08Rik |
RIKEN cDNA 4930458K08 gene |
1405 |
0.49 |
| chr1_162867938_162868165 | 0.28 |
Fmo1 |
flavin containing monooxygenase 1 |
1441 |
0.39 |
| chr14_9492266_9492441 | 0.28 |
Gm48371 |
predicted gene, 48371 |
22798 |
0.26 |
| chr7_132941355_132941506 | 0.28 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
6738 |
0.15 |
| chr2_31502991_31503411 | 0.27 |
Ass1 |
argininosuccinate synthetase 1 |
2475 |
0.27 |
| chr4_32561294_32561627 | 0.27 |
Bach2os |
BTB and CNC homology 2, opposite strand |
10157 |
0.16 |
| chr10_43441199_43441579 | 0.27 |
Gm3699 |
predicted gene 3699 |
15112 |
0.13 |
| chr2_75605802_75605972 | 0.27 |
Gm13655 |
predicted gene 13655 |
27495 |
0.15 |
| chr15_3463467_3463618 | 0.27 |
Ghr |
growth hormone receptor |
8102 |
0.29 |
| chr2_34049474_34049625 | 0.27 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr3_148832133_148832284 | 0.27 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
1720 |
0.51 |
| chr1_145995029_145995180 | 0.27 |
Gm5263 |
predicted gene 5263 |
425332 |
0.01 |
| chr2_71499002_71499251 | 0.27 |
Gm23253 |
predicted gene, 23253 |
3855 |
0.16 |
| chr1_167414950_167415113 | 0.27 |
Mgst3 |
microsomal glutathione S-transferase 3 |
21190 |
0.15 |
| chr6_59412258_59412432 | 0.27 |
Gprin3 |
GPRIN family member 3 |
13949 |
0.29 |
| chr14_26196215_26196366 | 0.26 |
Tmem254b |
transmembrane protein 254b |
8867 |
0.12 |
| chr1_85165585_85165793 | 0.26 |
Gm6264 |
predicted gene 6264 |
4826 |
0.12 |
| chr9_46479855_46480133 | 0.26 |
Gm47144 |
predicted gene, 47144 |
25079 |
0.15 |
| chr8_108703572_108703757 | 0.26 |
Zfhx3 |
zinc finger homeobox 3 |
564 |
0.82 |
| chr15_76269579_76269775 | 0.26 |
Mirt2 |
myocardial infraction associated transcript 2 |
52 |
0.92 |
| chr10_45048433_45048599 | 0.26 |
Prep |
prolyl endopeptidase |
18687 |
0.15 |
| chr19_31886956_31887132 | 0.26 |
A1cf |
APOBEC1 complementation factor |
18263 |
0.2 |
| chr19_56821183_56821529 | 0.25 |
Ccdc186 |
coiled-coil domain containing 186 |
834 |
0.58 |
| chr10_81407875_81408026 | 0.25 |
Nfic |
nuclear factor I/C |
309 |
0.68 |
| chr10_31594029_31594273 | 0.25 |
Rnf217 |
ring finger protein 217 |
15033 |
0.16 |
| chr4_101198122_101198415 | 0.25 |
Gm24468 |
predicted gene, 24468 |
11504 |
0.14 |
| chr8_46365811_46365999 | 0.25 |
Gm30931 |
predicted gene, 30931 |
2649 |
0.2 |
| chr8_36739901_36740056 | 0.25 |
Dlc1 |
deleted in liver cancer 1 |
6924 |
0.31 |
| chr4_80919092_80919518 | 0.25 |
Lurap1l |
leucine rich adaptor protein 1-like |
8659 |
0.26 |
| chr1_63136268_63136443 | 0.25 |
Ndufs1 |
NADH:ubiquinone oxidoreductase core subunit S1 |
8606 |
0.09 |
| chr18_61841168_61841343 | 0.25 |
Afap1l1 |
actin filament associated protein 1-like 1 |
54553 |
0.1 |
| chr1_23913075_23913318 | 0.25 |
Smap1 |
small ArfGAP 1 |
3460 |
0.31 |
| chr2_157131528_157131679 | 0.25 |
Samhd1 |
SAM domain and HD domain, 1 |
1546 |
0.33 |
| chr18_44633864_44634028 | 0.24 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
27719 |
0.19 |
| chr3_83048403_83048757 | 0.24 |
Fgb |
fibrinogen beta chain |
1283 |
0.37 |
| chr2_72309170_72309345 | 0.24 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
11356 |
0.19 |
| chr10_25563128_25563279 | 0.24 |
Gm29571 |
predicted gene 29571 |
26817 |
0.14 |
| chr5_102010214_102010423 | 0.24 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
28760 |
0.16 |
| chr15_36375228_36375396 | 0.24 |
Gm34150 |
predicted gene, 34150 |
263 |
0.88 |
| chr14_65375161_65375312 | 0.24 |
Zfp395 |
zinc finger protein 395 |
157 |
0.95 |
| chr10_69268049_69268226 | 0.24 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2500 |
0.29 |
| chr16_17069481_17069751 | 0.24 |
Ypel1 |
yippee like 1 |
80 |
0.93 |
| chr8_121898077_121898240 | 0.23 |
Slc7a5 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
1656 |
0.23 |
| chr17_29711245_29711660 | 0.23 |
Ccdc167 |
coiled-coil domain containing 167 |
5511 |
0.14 |
| chr15_64309233_64309403 | 0.23 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
2371 |
0.28 |
| chr6_138138988_138139150 | 0.23 |
Mgst1 |
microsomal glutathione S-transferase 1 |
1247 |
0.59 |
| chr4_57160943_57161117 | 0.23 |
Gm12530 |
predicted gene 12530 |
11041 |
0.2 |
| chr11_38683198_38683743 | 0.23 |
Gm23520 |
predicted gene, 23520 |
114807 |
0.07 |
| chr13_93629307_93629685 | 0.23 |
Gm15622 |
predicted gene 15622 |
4114 |
0.18 |
| chr10_62592617_62592774 | 0.23 |
Ddx21 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 |
1724 |
0.23 |
| chr1_127787130_127787281 | 0.23 |
Ccnt2 |
cyclin T2 |
9366 |
0.16 |
| chr4_124809297_124809476 | 0.23 |
Mtf1 |
metal response element binding transcription factor 1 |
6708 |
0.1 |
| chr19_39108442_39108835 | 0.23 |
Cyp2c66 |
cytochrome P450, family 2, subfamily c, polypeptide 66 |
5260 |
0.24 |
| chr19_24153611_24153973 | 0.23 |
Gm50308 |
predicted gene, 50308 |
278 |
0.9 |
| chr8_46908148_46908299 | 0.23 |
1700011L03Rik |
RIKEN cDNA 1700011L03 gene |
40576 |
0.13 |
| chr2_4895477_4895652 | 0.23 |
Sephs1 |
selenophosphate synthetase 1 |
11180 |
0.15 |
| chr4_55245568_55245866 | 0.23 |
Gm12508 |
predicted gene 12508 |
9023 |
0.17 |
| chr1_162813245_162813512 | 0.23 |
Fmo4 |
flavin containing monooxygenase 4 |
294 |
0.9 |
| chr7_75852856_75853007 | 0.23 |
Klhl25 |
kelch-like 25 |
4490 |
0.26 |
| chr2_118300879_118301335 | 0.23 |
1700054M17Rik |
RIKEN cDNA 1700054M17 gene |
3807 |
0.17 |
| chr2_132247965_132248291 | 0.23 |
Tmem230 |
transmembrane protein 230 |
321 |
0.83 |
| chr11_106889132_106889465 | 0.23 |
Smurf2 |
SMAD specific E3 ubiquitin protein ligase 2 |
30977 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.4 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.3 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.3 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.2 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.0 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.3 | GO:0044688 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |