Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
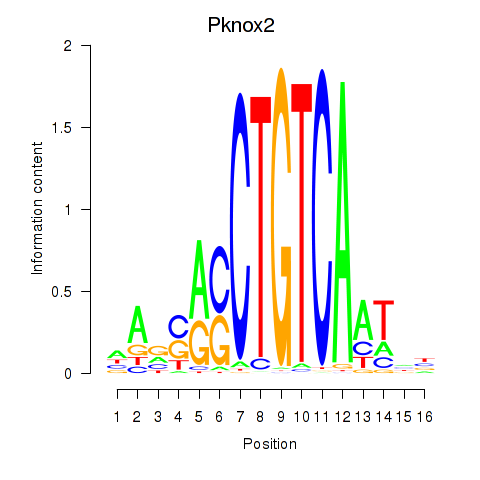
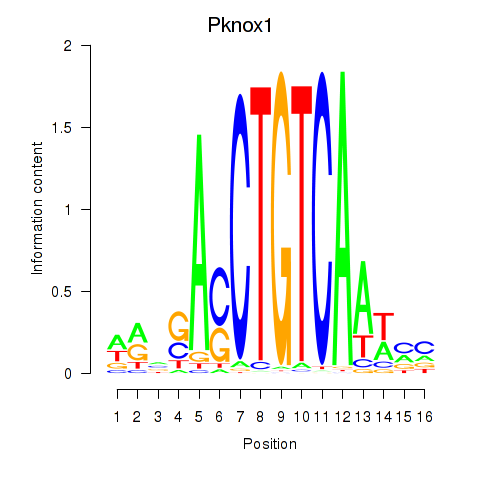
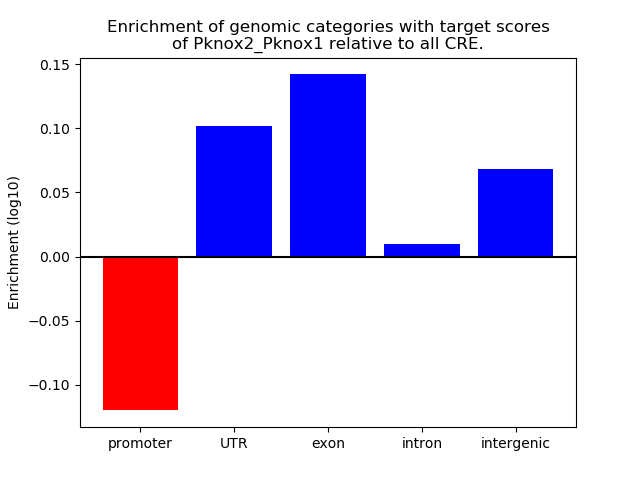
Results for Pknox2_Pknox1
Z-value: 0.95
Transcription factors associated with Pknox2_Pknox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pknox2
|
ENSMUSG00000035934.9 | Pbx/knotted 1 homeobox 2 |
|
Pknox1
|
ENSMUSG00000006705.6 | Pbx/knotted 1 homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_31607362_31607553 | Pknox1 | 4719 | 0.101046 | 0.91 | 1.1e-02 | Click! |
| chr17_31607662_31607820 | Pknox1 | 5003 | 0.099351 | 0.79 | 6.4e-02 | Click! |
| chr17_31608379_31608531 | Pknox1 | 5717 | 0.096085 | 0.59 | 2.1e-01 | Click! |
| chr17_31564751_31564906 | Pknox1 | 12 | 0.695950 | 0.58 | 2.2e-01 | Click! |
| chr17_31607121_31607285 | Pknox1 | 4465 | 0.102826 | 0.40 | 4.3e-01 | Click! |
| chr9_37147174_37147330 | Pknox2 | 14 | 0.964661 | 0.81 | 4.9e-02 | Click! |
| chr9_36933794_36933960 | Pknox2 | 53395 | 0.099241 | -0.77 | 7.5e-02 | Click! |
| chr9_36991937_36992116 | Pknox2 | 4754 | 0.192169 | 0.71 | 1.2e-01 | Click! |
| chr9_37146676_37146845 | Pknox2 | 506 | 0.701499 | 0.70 | 1.2e-01 | Click! |
| chr9_37146437_37146602 | Pknox2 | 704 | 0.570203 | 0.62 | 1.9e-01 | Click! |
Activity of the Pknox2_Pknox1 motif across conditions
Conditions sorted by the z-value of the Pknox2_Pknox1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
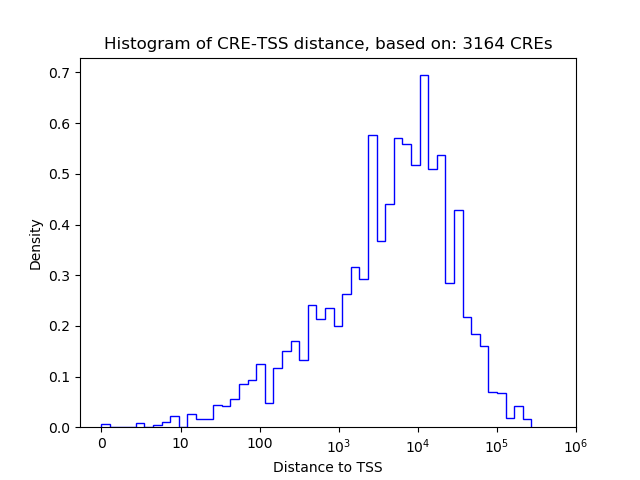
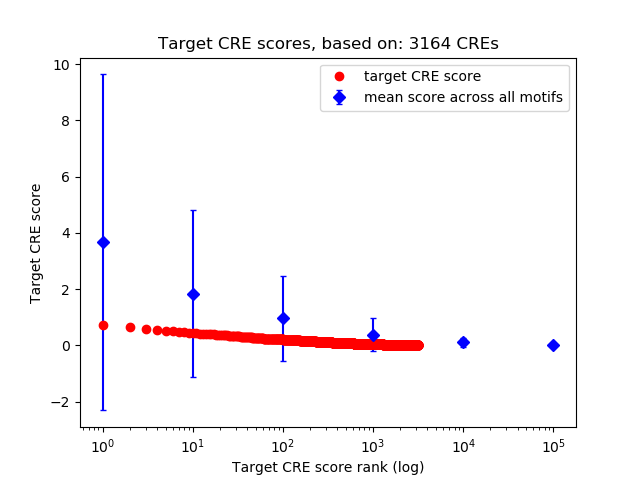
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_180219669_180219845 | 0.72 |
Gm37336 |
predicted gene, 37336 |
7356 |
0.14 |
| chr10_119010707_119010858 | 0.63 |
Gm47461 |
predicted gene, 47461 |
10888 |
0.17 |
| chr12_21166146_21166319 | 0.60 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
54278 |
0.1 |
| chr6_121860064_121860215 | 0.53 |
Mug1 |
murinoglobulin 1 |
19035 |
0.19 |
| chr4_60659551_60660018 | 0.50 |
Mup11 |
major urinary protein 11 |
46 |
0.97 |
| chr9_69306275_69306692 | 0.50 |
Rora |
RAR-related orphan receptor alpha |
16801 |
0.21 |
| chr12_40475800_40475951 | 0.47 |
Dock4 |
dedicator of cytokinesis 4 |
29539 |
0.19 |
| chr2_19647255_19647467 | 0.47 |
Gm13343 |
predicted gene 13343 |
4987 |
0.15 |
| chr15_31058451_31058629 | 0.43 |
4930430F21Rik |
RIKEN cDNA 4930430F21 gene |
19412 |
0.2 |
| chr1_93146606_93146776 | 0.43 |
Agxt |
alanine-glyoxylate aminotransferase |
6812 |
0.13 |
| chr4_33245665_33245911 | 0.43 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
1787 |
0.31 |
| chr18_46717220_46717635 | 0.42 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
10602 |
0.13 |
| chr12_109993740_109993939 | 0.40 |
Gm34667 |
predicted gene, 34667 |
30034 |
0.1 |
| chr12_12405687_12405864 | 0.40 |
4921511I17Rik |
RIKEN cDNA 4921511I17 gene |
13160 |
0.28 |
| chr10_78294277_78294441 | 0.40 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
1035 |
0.32 |
| chr1_191493887_191494044 | 0.39 |
Gm37432 |
predicted gene, 37432 |
60 |
0.96 |
| chr7_113223800_113224075 | 0.39 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
10686 |
0.21 |
| chr9_50875623_50875774 | 0.38 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
2387 |
0.25 |
| chr19_23060777_23060935 | 0.37 |
Gm50136 |
predicted gene, 50136 |
598 |
0.75 |
| chr19_36627452_36627789 | 0.37 |
Hectd2os |
Hectd2, opposite strand |
1596 |
0.43 |
| chr17_56211204_56211384 | 0.36 |
Dpp9 |
dipeptidylpeptidase 9 |
2086 |
0.16 |
| chr9_48739409_48739562 | 0.36 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
96460 |
0.07 |
| chr10_63410268_63410432 | 0.36 |
Gm7530 |
predicted gene 7530 |
2649 |
0.19 |
| chr1_93687795_93687979 | 0.35 |
Bok |
BCL2-related ovarian killer |
1321 |
0.35 |
| chr10_43709388_43709808 | 0.35 |
F930017D23Rik |
RIKEN cDNA F930017D23 gene |
2700 |
0.18 |
| chr13_93632435_93632697 | 0.34 |
Bhmt |
betaine-homocysteine methyltransferase |
5000 |
0.17 |
| chr16_35727863_35728078 | 0.34 |
Gm25967 |
predicted gene, 25967 |
5737 |
0.18 |
| chr9_40638323_40638501 | 0.34 |
Gm48284 |
predicted gene, 48284 |
22026 |
0.1 |
| chr5_114553798_114553965 | 0.33 |
Gm13790 |
predicted gene 13790 |
2592 |
0.24 |
| chr16_16225035_16225410 | 0.33 |
Pkp2 |
plakophilin 2 |
11904 |
0.19 |
| chr14_21719739_21719901 | 0.33 |
Dupd1 |
dual specificity phosphatase and pro isomerase domain containing 1 |
5244 |
0.17 |
| chr5_114553572_114553723 | 0.33 |
Gm13790 |
predicted gene 13790 |
2358 |
0.25 |
| chr11_46312714_46312865 | 0.32 |
Cyfip2 |
cytoplasmic FMR1 interacting protein 2 |
70 |
0.97 |
| chr1_190044023_190044190 | 0.31 |
Smyd2 |
SET and MYND domain containing 2 |
121743 |
0.05 |
| chr15_67420967_67421299 | 0.31 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
194364 |
0.03 |
| chr16_94693543_94693992 | 0.31 |
Gm41504 |
predicted gene, 41504 |
20136 |
0.16 |
| chr15_80680125_80680365 | 0.30 |
Fam83f |
family with sequence similarity 83, member F |
8398 |
0.13 |
| chr11_102753302_102753453 | 0.30 |
Adam11 |
a disintegrin and metallopeptidase domain 11 |
8062 |
0.11 |
| chr9_66154137_66154300 | 0.29 |
Dapk2 |
death-associated protein kinase 2 |
4005 |
0.21 |
| chr5_130761960_130762123 | 0.29 |
Gm23761 |
predicted gene, 23761 |
20660 |
0.24 |
| chr16_93728444_93728595 | 0.29 |
Dop1b |
DOP1 leucine zipper like protein B |
1580 |
0.35 |
| chr13_112071667_112072207 | 0.28 |
Gm31104 |
predicted gene, 31104 |
66179 |
0.09 |
| chr2_164054518_164054669 | 0.28 |
Pabpc1l |
poly(A) binding protein, cytoplasmic 1-like |
6583 |
0.14 |
| chr3_132887148_132887306 | 0.28 |
Gm29811 |
predicted gene, 29811 |
8631 |
0.16 |
| chr4_107002621_107002790 | 0.28 |
Gm12786 |
predicted gene 12786 |
19794 |
0.15 |
| chr9_70931148_70931345 | 0.27 |
Gm32017 |
predicted gene, 32017 |
758 |
0.66 |
| chr19_36484338_36484507 | 0.27 |
F530104D19Rik |
RIKEN cDNA F530104D19 gene |
33265 |
0.15 |
| chr8_119428106_119428304 | 0.27 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
5919 |
0.16 |
| chr1_121304990_121305141 | 0.26 |
Insig2 |
insulin induced gene 2 |
2253 |
0.26 |
| chr15_37456125_37456321 | 0.26 |
Ncald |
neurocalcin delta |
2318 |
0.22 |
| chr3_83043842_83044694 | 0.26 |
Fgb |
fibrinogen beta chain |
5595 |
0.15 |
| chr3_8891989_8892146 | 0.26 |
Gm15466 |
predicted gene 15466 |
9497 |
0.17 |
| chr7_79493997_79494159 | 0.26 |
Mir9-3hg |
Mir9-3 host gene |
5948 |
0.1 |
| chr4_137481328_137481504 | 0.26 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
12613 |
0.13 |
| chr15_102192070_102192387 | 0.25 |
Csad |
cysteine sulfinic acid decarboxylase |
169 |
0.91 |
| chr4_45285469_45285620 | 0.25 |
Trmt10b |
tRNA methyltransferase 10B |
11583 |
0.15 |
| chr10_8089529_8089695 | 0.25 |
Gm48614 |
predicted gene, 48614 |
68320 |
0.11 |
| chr8_48729447_48729608 | 0.25 |
Tenm3 |
teneurin transmembrane protein 3 |
54837 |
0.15 |
| chr15_36471981_36472313 | 0.25 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr3_81956990_81957439 | 0.24 |
Ctso |
cathepsin O |
6277 |
0.16 |
| chr11_58003210_58003371 | 0.24 |
Larp1 |
La ribonucleoprotein domain family, member 1 |
5774 |
0.17 |
| chr4_61436878_61437320 | 0.24 |
Mup15 |
major urinary protein 15 |
2644 |
0.27 |
| chr7_125577330_125577481 | 0.24 |
Gm44876 |
predicted gene 44876 |
5288 |
0.17 |
| chr11_46579639_46579790 | 0.24 |
BC053393 |
cDNA sequence BC053393 |
8178 |
0.13 |
| chr19_3840335_3840486 | 0.24 |
Chka |
choline kinase alpha |
11363 |
0.09 |
| chr3_104796399_104796698 | 0.23 |
Rhoc |
ras homolog family member C |
4565 |
0.1 |
| chr5_114982798_114982985 | 0.23 |
1810017P11Rik |
RIKEN cDNA 1810017P11 gene |
7435 |
0.09 |
| chr15_78231969_78232141 | 0.23 |
Ncf4 |
neutrophil cytosolic factor 4 |
12746 |
0.13 |
| chr6_91686720_91686890 | 0.23 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
2656 |
0.18 |
| chr9_20504907_20505058 | 0.23 |
Zfp426 |
zinc finger protein 426 |
12236 |
0.11 |
| chr10_34298110_34298293 | 0.23 |
Gm47512 |
predicted gene, 47512 |
613 |
0.44 |
| chr3_50443598_50443859 | 0.22 |
Slc7a11 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
114 |
0.97 |
| chr19_61049084_61049293 | 0.22 |
Gm22520 |
predicted gene, 22520 |
35643 |
0.14 |
| chr10_86069565_86069729 | 0.22 |
Syn3 |
synapsin III |
2804 |
0.18 |
| chr17_84895942_84896103 | 0.22 |
Gm49982 |
predicted gene, 49982 |
6547 |
0.18 |
| chr8_86933808_86934280 | 0.22 |
Gm27168 |
predicted gene 27168 |
12897 |
0.13 |
| chr9_74868478_74868651 | 0.22 |
Onecut1 |
one cut domain, family member 1 |
2080 |
0.26 |
| chr1_179844970_179845129 | 0.22 |
Ahctf1 |
AT hook containing transcription factor 1 |
41369 |
0.14 |
| chr7_114203119_114203440 | 0.22 |
Gm45454 |
predicted gene 45454 |
5809 |
0.22 |
| chr3_60057385_60057653 | 0.22 |
Sucnr1 |
succinate receptor 1 |
24350 |
0.14 |
| chr17_13699350_13699759 | 0.22 |
Gm16046 |
predicted gene 16046 |
16038 |
0.13 |
| chr6_93153422_93153582 | 0.22 |
Gm5313 |
predicted gene 5313 |
15512 |
0.21 |
| chr4_61779591_61780024 | 0.22 |
Mup19 |
major urinary protein 19 |
2417 |
0.19 |
| chr17_64609042_64609223 | 0.22 |
Man2a1 |
mannosidase 2, alpha 1 |
8396 |
0.26 |
| chr9_65322219_65322370 | 0.22 |
Gm39363 |
predicted gene, 39363 |
10226 |
0.09 |
| chr14_66672460_66672626 | 0.22 |
Adra1a |
adrenergic receptor, alpha 1a |
36987 |
0.17 |
| chr9_119143465_119143650 | 0.22 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
6506 |
0.12 |
| chr9_63042658_63043014 | 0.22 |
Gm24526 |
predicted gene, 24526 |
15276 |
0.18 |
| chr4_61671437_61671659 | 0.22 |
Mup18 |
major urinary protein 18 |
2588 |
0.25 |
| chr2_101903298_101903465 | 0.22 |
Commd9 |
COMM domain containing 9 |
17118 |
0.18 |
| chr1_5071962_5072114 | 0.22 |
Rgs20 |
regulator of G-protein signaling 20 |
1753 |
0.28 |
| chr19_12486979_12487160 | 0.21 |
Dtx4 |
deltex 4, E3 ubiquitin ligase |
14385 |
0.1 |
| chr17_50489431_50489587 | 0.21 |
Plcl2 |
phospholipase C-like 2 |
19894 |
0.26 |
| chr15_102186991_102187189 | 0.21 |
Csad |
cysteine sulfinic acid decarboxylase |
509 |
0.65 |
| chr7_48881274_48881434 | 0.21 |
Gm2788 |
predicted gene 2788 |
54 |
0.74 |
| chr4_19681355_19681795 | 0.21 |
Wwp1 |
WW domain containing E3 ubiquitin protein ligase 1 |
3127 |
0.29 |
| chr10_83112464_83112627 | 0.21 |
1700025N21Rik |
RIKEN cDNA 1700025N21 gene |
29493 |
0.17 |
| chr19_25542114_25542287 | 0.21 |
Dmrt1 |
doublesex and mab-3 related transcription factor 1 |
36493 |
0.15 |
| chr2_116065184_116065343 | 0.21 |
Meis2 |
Meis homeobox 2 |
216 |
0.9 |
| chr9_111100529_111100680 | 0.21 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
16988 |
0.12 |
| chr19_47222639_47222810 | 0.21 |
Gm50339 |
predicted gene, 50339 |
815 |
0.48 |
| chr8_117700284_117700470 | 0.20 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
1527 |
0.3 |
| chr11_76709264_76709415 | 0.20 |
Trarg1 |
trafficking regulator of GLUT4 (SLC2A4) 1 |
29531 |
0.14 |
| chr2_132577301_132577957 | 0.20 |
Gpcpd1 |
glycerophosphocholine phosphodiesterase 1 |
504 |
0.77 |
| chr6_94550696_94550886 | 0.20 |
Slc25a26 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
16651 |
0.2 |
| chr2_122944888_122945077 | 0.20 |
Sqor |
sulfide quinone oxidoreductase |
145220 |
0.04 |
| chr15_86177571_86177722 | 0.20 |
Gm15569 |
predicted gene 15569 |
8082 |
0.17 |
| chr1_37300588_37300990 | 0.19 |
Inpp4a |
inositol polyphosphate-4-phosphatase, type I |
481 |
0.66 |
| chr2_169750622_169750798 | 0.19 |
Tshz2 |
teashirt zinc finger family member 2 |
117034 |
0.06 |
| chr9_96977502_96977653 | 0.19 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
5602 |
0.18 |
| chr8_126806262_126806425 | 0.19 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
32890 |
0.16 |
| chr5_66798803_66798961 | 0.19 |
Limch1 |
LIM and calponin homology domains 1 |
52993 |
0.11 |
| chr9_100446507_100446671 | 0.19 |
Gm28166 |
predicted gene 28166 |
3733 |
0.19 |
| chr8_119436291_119436442 | 0.19 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
752 |
0.59 |
| chr3_81062807_81062975 | 0.19 |
Gm16000 |
predicted gene 16000 |
22454 |
0.16 |
| chr8_10965649_10965912 | 0.19 |
Gm44956 |
predicted gene 44956 |
5034 |
0.13 |
| chr5_100860430_100860646 | 0.19 |
4930458D05Rik |
RIKEN cDNA 4930458D05 gene |
13371 |
0.1 |
| chr3_85821665_85821854 | 0.19 |
Fam160a1 |
family with sequence similarity 160, member A1 |
4468 |
0.2 |
| chr18_46627491_46627728 | 0.19 |
Gm3734 |
predicted gene 3734 |
3198 |
0.2 |
| chr19_36534453_36534632 | 0.19 |
Hectd2 |
HECT domain E3 ubiquitin protein ligase 2 |
20097 |
0.2 |
| chr2_6126122_6126290 | 0.19 |
Proser2 |
proline and serine rich 2 |
3933 |
0.18 |
| chr19_47410387_47410546 | 0.19 |
Sh3pxd2a |
SH3 and PX domains 2A |
107 |
0.97 |
| chr1_171196191_171196374 | 0.19 |
Pcp4l1 |
Purkinje cell protein 4-like 1 |
14 |
0.94 |
| chr14_22800645_22800832 | 0.19 |
Gm7473 |
predicted gene 7473 |
25494 |
0.25 |
| chr4_60067672_60068037 | 0.19 |
Mup7 |
major urinary protein 7 |
2557 |
0.26 |
| chr2_32389286_32389437 | 0.19 |
Lcn2 |
lipocalin 2 |
1109 |
0.29 |
| chr9_66331624_66331801 | 0.18 |
Herc1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
18738 |
0.18 |
| chr15_99867784_99867962 | 0.18 |
Lima1 |
LIM domain and actin binding 1 |
6798 |
0.08 |
| chr1_67038526_67038707 | 0.18 |
Lancl1 |
LanC (bacterial lantibiotic synthetase component C)-like 1 |
218 |
0.94 |
| chr6_122598952_122599267 | 0.18 |
Apobec1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
1018 |
0.37 |
| chr15_86202799_86202950 | 0.18 |
Gm22818 |
predicted gene, 22818 |
9783 |
0.16 |
| chr4_144961736_144961902 | 0.18 |
Gm38074 |
predicted gene, 38074 |
2971 |
0.26 |
| chr4_149794929_149795189 | 0.18 |
Gm13065 |
predicted gene 13065 |
1640 |
0.21 |
| chr1_93124444_93124635 | 0.18 |
Gm28086 |
predicted gene 28086 |
436 |
0.76 |
| chr8_117736689_117736847 | 0.18 |
Gm31774 |
predicted gene, 31774 |
11492 |
0.14 |
| chr19_36731141_36731447 | 0.18 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
5359 |
0.23 |
| chr2_28525746_28525944 | 0.18 |
Ralgds |
ral guanine nucleotide dissociation stimulator |
2005 |
0.19 |
| chr2_167982734_167983076 | 0.18 |
Ripor3 |
RIPOR family member 3 |
3850 |
0.2 |
| chr5_103591965_103592170 | 0.18 |
Gm15844 |
predicted gene 15844 |
32404 |
0.12 |
| chr4_62778005_62778291 | 0.18 |
Gm24117 |
predicted gene, 24117 |
34280 |
0.14 |
| chr11_83852040_83852214 | 0.18 |
Hnf1b |
HNF1 homeobox B |
115 |
0.95 |
| chr18_68005512_68005663 | 0.18 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
72330 |
0.09 |
| chr1_132170141_132170558 | 0.18 |
Lemd1 |
LEM domain containing 1 |
21082 |
0.1 |
| chr10_86092029_86092180 | 0.18 |
4930486F22Rik |
RIKEN cDNA 4930486F22 gene |
3755 |
0.17 |
| chr8_60631719_60632037 | 0.18 |
Gm34730 |
predicted gene, 34730 |
853 |
0.44 |
| chr17_35251426_35251588 | 0.18 |
Mir8094 |
microRNA 8094 |
218 |
0.76 |
| chr4_60137209_60137437 | 0.17 |
Mup2 |
major urinary protein 2 |
2534 |
0.26 |
| chr4_97988281_97988451 | 0.17 |
Nfia |
nuclear factor I/A |
77333 |
0.11 |
| chrX_36874176_36874336 | 0.17 |
Ube2a |
ubiquitin-conjugating enzyme E2A |
39 |
0.63 |
| chr10_76032130_76032292 | 0.17 |
Zfp280b |
zinc finger protein 280B |
439 |
0.73 |
| chr2_43678933_43679111 | 0.17 |
Kynu |
kynureninase |
8092 |
0.3 |
| chr5_130853198_130853625 | 0.17 |
Gm42894 |
predicted gene 42894 |
14426 |
0.22 |
| chr8_101351773_101352122 | 0.17 |
Gm22223 |
predicted gene, 22223 |
187761 |
0.03 |
| chr3_27788248_27788425 | 0.17 |
Fndc3b |
fibronectin type III domain containing 3B |
77029 |
0.1 |
| chr8_21778650_21778989 | 0.17 |
Defb1 |
defensin beta 1 |
2220 |
0.16 |
| chr4_118132141_118132338 | 0.17 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
2643 |
0.23 |
| chr16_23106926_23107120 | 0.17 |
Eif4a2 |
eukaryotic translation initiation factor 4A2 |
421 |
0.55 |
| chr8_33881292_33881487 | 0.17 |
Gm26978 |
predicted gene, 26978 |
4358 |
0.2 |
| chr2_163494018_163494324 | 0.17 |
R3hdml |
R3H domain containing-like |
1853 |
0.21 |
| chr11_78496004_78496169 | 0.17 |
Sarm1 |
sterile alpha and HEAT/Armadillo motif containing 1 |
1372 |
0.2 |
| chr11_78417415_78417566 | 0.17 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
4727 |
0.11 |
| chr13_45906518_45907031 | 0.17 |
4930453C13Rik |
RIKEN cDNA 4930453C13 gene |
29266 |
0.18 |
| chr16_57168528_57168697 | 0.16 |
Gm16892 |
predicted gene, 16892 |
1211 |
0.31 |
| chr7_141949547_141949705 | 0.16 |
Gm20501 |
predicted gene 20501 |
87 |
0.55 |
| chr5_5211241_5211412 | 0.16 |
Cdk14 |
cyclin-dependent kinase 14 |
15971 |
0.18 |
| chr3_51560826_51560999 | 0.16 |
Setd7 |
SET domain containing (lysine methyltransferase) 7 |
33 |
0.92 |
| chr15_55224563_55224882 | 0.16 |
Gm26296 |
predicted gene, 26296 |
37881 |
0.16 |
| chr19_57535215_57535466 | 0.16 |
Gm50279 |
predicted gene, 50279 |
5443 |
0.15 |
| chr2_30957612_30957790 | 0.16 |
Tor1b |
torsin family 1, member B |
2448 |
0.18 |
| chr19_42196519_42196698 | 0.16 |
Sfrp5 |
secreted frizzled-related sequence protein 5 |
5644 |
0.14 |
| chr6_121892053_121892211 | 0.16 |
Mug1 |
murinoglobulin 1 |
6555 |
0.19 |
| chr10_77064998_77065227 | 0.16 |
Col18a1 |
collagen, type XVIII, alpha 1 |
5115 |
0.17 |
| chrX_102478577_102478728 | 0.16 |
Hdac8 |
histone deacetylase 8 |
26089 |
0.19 |
| chr9_108794998_108795174 | 0.16 |
Gm23034 |
predicted gene, 23034 |
302 |
0.72 |
| chr17_29053774_29054133 | 0.16 |
Gm41556 |
predicted gene, 41556 |
154 |
0.9 |
| chr7_30353715_30354115 | 0.16 |
4930479H17Rik |
RIKEN cDNA 4930479H17 gene |
594 |
0.46 |
| chr8_117306309_117306592 | 0.16 |
Cmip |
c-Maf inducing protein |
42720 |
0.17 |
| chr10_115777494_115777663 | 0.16 |
Tspan8 |
tetraspanin 8 |
39254 |
0.19 |
| chr11_90258619_90258781 | 0.16 |
Mmd |
monocyte to macrophage differentiation-associated |
9224 |
0.22 |
| chr4_60818855_60819228 | 0.16 |
Mup22 |
major urinary protein 22 |
2422 |
0.29 |
| chr16_10952104_10952327 | 0.15 |
Gm26268 |
predicted gene, 26268 |
13509 |
0.11 |
| chr5_145984724_145985158 | 0.15 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
6702 |
0.13 |
| chr12_104818327_104818478 | 0.15 |
Clmn |
calmin |
37531 |
0.14 |
| chr10_99599315_99599605 | 0.15 |
Gm20110 |
predicted gene, 20110 |
9711 |
0.18 |
| chr15_100659832_100659991 | 0.15 |
Bin2 |
bridging integrator 2 |
8251 |
0.09 |
| chr7_141475336_141475516 | 0.15 |
Tspan4 |
tetraspanin 4 |
84 |
0.73 |
| chr1_39105102_39105502 | 0.15 |
Gm37091 |
predicted gene, 37091 |
21964 |
0.17 |
| chr10_127877485_127877659 | 0.15 |
Rdh7 |
retinol dehydrogenase 7 |
10758 |
0.09 |
| chr8_10859650_10859809 | 0.15 |
Gm32540 |
predicted gene, 32540 |
6457 |
0.13 |
| chr11_20831246_20831415 | 0.15 |
Lgalsl |
lectin, galactoside binding-like |
274 |
0.72 |
| chr1_151679282_151679446 | 0.15 |
Fam129a |
family with sequence similarity 129, member A |
2197 |
0.35 |
| chr14_29107334_29107489 | 0.15 |
Cacna2d3 |
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
9508 |
0.21 |
| chr5_43219459_43219610 | 0.15 |
Cpeb2 |
cytoplasmic polyadenylation element binding protein 2 |
13636 |
0.15 |
| chr19_45745506_45745808 | 0.15 |
Fgf8 |
fibroblast growth factor 8 |
2742 |
0.18 |
| chr19_34880110_34880491 | 0.15 |
Pank1 |
pantothenate kinase 1 |
845 |
0.59 |
| chr15_36491785_36492199 | 0.15 |
Ankrd46 |
ankyrin repeat domain 46 |
4723 |
0.17 |
| chr13_35018660_35018815 | 0.15 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
8357 |
0.12 |
| chr19_55660221_55660414 | 0.15 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
81503 |
0.11 |
| chr14_41119395_41119820 | 0.15 |
Sftpa1 |
surfactant associated protein A1 |
12175 |
0.11 |
| chr13_36577900_36578089 | 0.14 |
Gm48767 |
predicted gene, 48767 |
2060 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.0 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.0 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.0 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.0 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |