Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
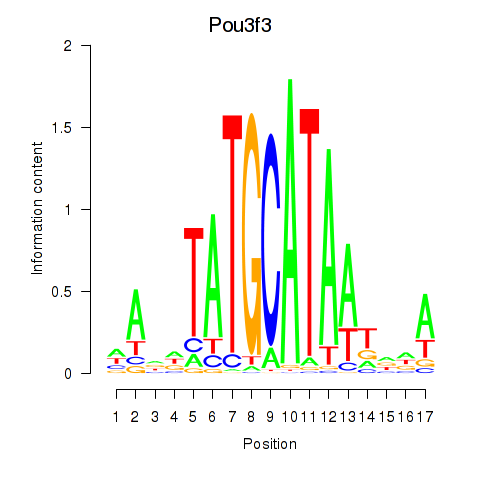
Results for Pou3f3
Z-value: 1.00
Transcription factors associated with Pou3f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f3
|
ENSMUSG00000045515.2 | POU domain, class 3, transcription factor 3 |
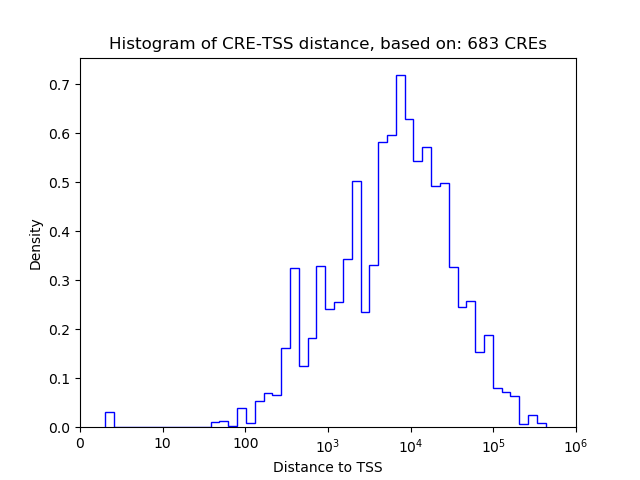
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_42699734_42699893 | Pou3f3 | 4045 | 0.155167 | 0.85 | 3.0e-02 | Click! |
Activity of the Pou3f3 motif across conditions
Conditions sorted by the z-value of the Pou3f3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
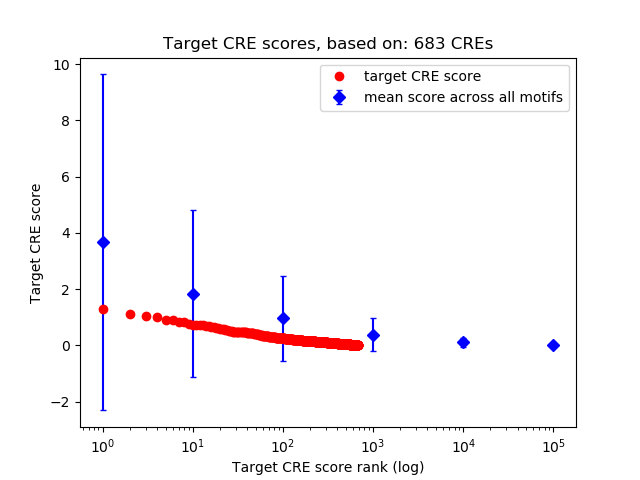
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_3046150_3046544 | 1.29 |
Gm44634 |
predicted gene 44634 |
9947 |
0.2 |
| chr3_51255928_51256079 | 1.10 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr10_89492629_89492900 | 1.03 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
13885 |
0.21 |
| chr3_97649987_97650151 | 1.01 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
8124 |
0.13 |
| chr13_43549369_43549520 | 0.91 |
Mcur1 |
mitochondrial calcium uniporter regulator 1 |
3324 |
0.2 |
| chr3_144109828_144110050 | 0.89 |
Gm34078 |
predicted gene, 34078 |
25815 |
0.2 |
| chr15_3470264_3470480 | 0.84 |
Ghr |
growth hormone receptor |
1272 |
0.6 |
| chr8_93183570_93183871 | 0.84 |
Gm45909 |
predicted gene 45909 |
7638 |
0.14 |
| chr16_37902225_37902391 | 0.74 |
Gpr156 |
G protein-coupled receptor 156 |
14188 |
0.14 |
| chr8_93164567_93164980 | 0.73 |
Ces1d |
carboxylesterase 1D |
5202 |
0.15 |
| chr8_93185812_93185963 | 0.72 |
Gm45909 |
predicted gene 45909 |
5471 |
0.15 |
| chr9_70446864_70447026 | 0.71 |
Rnf111 |
ring finger 111 |
6901 |
0.16 |
| chr15_36280810_36280963 | 0.70 |
Rnf19a |
ring finger protein 19A |
2212 |
0.2 |
| chr1_67181891_67182275 | 0.70 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
59057 |
0.11 |
| chr1_67183580_67183935 | 0.68 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
60731 |
0.11 |
| chr11_11931538_11931698 | 0.65 |
Grb10 |
growth factor receptor bound protein 10 |
6998 |
0.19 |
| chr8_93165222_93165456 | 0.65 |
Ces1d |
carboxylesterase 1D |
4636 |
0.15 |
| chr1_31097093_31097258 | 0.63 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
721 |
0.62 |
| chr17_64676171_64676533 | 0.62 |
Man2a1 |
mannosidase 2, alpha 1 |
37068 |
0.18 |
| chr1_71723212_71723548 | 0.59 |
Gm5256 |
predicted gene 5256 |
8456 |
0.19 |
| chr2_58773911_58774203 | 0.57 |
Upp2 |
uridine phosphorylase 2 |
8732 |
0.21 |
| chr4_150866491_150867250 | 0.57 |
Errfi1 |
ERBB receptor feedback inhibitor 1 |
11797 |
0.13 |
| chr12_16633839_16633990 | 0.56 |
Lpin1 |
lipin 1 |
13052 |
0.17 |
| chr2_134947281_134947437 | 0.55 |
Gm14036 |
predicted gene 14036 |
143410 |
0.04 |
| chr6_13654409_13654594 | 0.52 |
Bmt2 |
base methyltransferase of 25S rRNA 2 |
23437 |
0.21 |
| chr11_31675803_31675997 | 0.52 |
Bod1 |
biorientation of chromosomes in cell division 1 |
4015 |
0.27 |
| chr1_106630171_106630322 | 0.51 |
Gm37053 |
predicted gene, 37053 |
23376 |
0.19 |
| chr5_8842744_8842895 | 0.49 |
Abcb1b |
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
2458 |
0.24 |
| chr11_5347339_5347533 | 0.48 |
Znrf3 |
zinc and ring finger 3 |
34304 |
0.15 |
| chr2_67977965_67978321 | 0.48 |
Gm37964 |
predicted gene, 37964 |
79547 |
0.1 |
| chr10_76531927_76532170 | 0.48 |
Lss |
lanosterol synthase |
366 |
0.79 |
| chr17_13821264_13821648 | 0.48 |
Afdn |
afadin, adherens junction formation factor |
29506 |
0.14 |
| chr6_6206187_6206411 | 0.47 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
10819 |
0.23 |
| chr8_93258499_93258650 | 0.47 |
Ces1f |
carboxylesterase 1F |
165 |
0.94 |
| chr6_138152720_138152908 | 0.47 |
Mgst1 |
microsomal glutathione S-transferase 1 |
9960 |
0.28 |
| chr1_13670589_13670740 | 0.47 |
Xkr9 |
X-linked Kx blood group related 9 |
1893 |
0.32 |
| chr11_103696519_103696670 | 0.47 |
Gosr2 |
golgi SNAP receptor complex member 2 |
1100 |
0.31 |
| chr4_35533897_35534048 | 0.47 |
Gm12365 |
predicted gene 12365 |
97758 |
0.08 |
| chr8_101350931_101351097 | 0.46 |
Gm22223 |
predicted gene, 22223 |
188694 |
0.03 |
| chr4_108085090_108085394 | 0.45 |
Mir6397 |
microRNA 6397 |
2115 |
0.24 |
| chr16_43286493_43286831 | 0.45 |
Gm37946 |
predicted gene, 37946 |
21619 |
0.16 |
| chr5_147726852_147727014 | 0.45 |
Flt1 |
FMS-like tyrosine kinase 1 |
922 |
0.61 |
| chr6_95652369_95652586 | 0.44 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
4991 |
0.35 |
| chr3_60097884_60098081 | 0.44 |
Sucnr1 |
succinate receptor 1 |
16080 |
0.18 |
| chr1_162892140_162892544 | 0.43 |
Fmo2 |
flavin containing monooxygenase 2 |
5827 |
0.19 |
| chr2_112284742_112285127 | 0.42 |
Slc12a6 |
solute carrier family 12, member 6 |
357 |
0.81 |
| chr16_42910018_42910191 | 0.42 |
Gm19522 |
predicted gene, 19522 |
1961 |
0.29 |
| chr19_17506726_17506895 | 0.42 |
Rfk |
riboflavin kinase |
109373 |
0.07 |
| chr16_56714341_56714503 | 0.42 |
Tfg |
Trk-fused gene |
1446 |
0.47 |
| chr3_151831850_151832001 | 0.41 |
Ptgfr |
prostaglandin F receptor |
2230 |
0.27 |
| chr19_40512528_40512992 | 0.39 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
913 |
0.57 |
| chr9_95558523_95558904 | 0.39 |
Paqr9 |
progestin and adipoQ receptor family member IX |
944 |
0.41 |
| chr13_102570134_102570306 | 0.39 |
Gm29927 |
predicted gene, 29927 |
20065 |
0.22 |
| chr5_147747897_147748048 | 0.38 |
Gm43156 |
predicted gene 43156 |
2456 |
0.3 |
| chr5_17964323_17964474 | 0.38 |
Gnat3 |
guanine nucleotide binding protein, alpha transducing 3 |
1849 |
0.49 |
| chr19_58049317_58049501 | 0.36 |
Mir5623 |
microRNA 5623 |
1758 |
0.46 |
| chr17_13825284_13825458 | 0.36 |
Afdn |
afadin, adherens junction formation factor |
25591 |
0.15 |
| chr8_20561636_20561787 | 0.36 |
Gm21182 |
predicted gene, 21182 |
4225 |
0.2 |
| chr8_25750286_25750467 | 0.35 |
Ddhd2 |
DDHD domain containing 2 |
526 |
0.61 |
| chr6_43643507_43643672 | 0.35 |
Tpk1 |
thiamine pyrophosphokinase |
17623 |
0.25 |
| chr12_4455371_4455763 | 0.34 |
Ncoa1 |
nuclear receptor coactivator 1 |
21615 |
0.16 |
| chr7_25422600_25422802 | 0.34 |
Gm20949 |
predicted gene, 20949 |
91 |
0.93 |
| chr7_26817080_26817232 | 0.34 |
Cyp2g1 |
cytochrome P450, family 2, subfamily g, polypeptide 1 |
861 |
0.56 |
| chr4_48225504_48225656 | 0.34 |
Mir1958 |
microRNA 1958 |
12365 |
0.19 |
| chr2_105949709_105950077 | 0.33 |
Gm22002 |
predicted gene, 22002 |
42864 |
0.1 |
| chr6_100858577_100858772 | 0.33 |
Ppp4r2 |
protein phosphatase 4, regulatory subunit 2 |
6097 |
0.23 |
| chr18_38370754_38370932 | 0.33 |
Gm4949 |
predicted gene 4949 |
10305 |
0.12 |
| chr11_54003633_54003801 | 0.33 |
Slc22a4 |
solute carrier family 22 (organic cation transporter), member 4 |
4247 |
0.16 |
| chr11_16780614_16780813 | 0.32 |
Egfr |
epidermal growth factor receptor |
28483 |
0.16 |
| chr13_7307728_7307879 | 0.31 |
Gm8725 |
predicted gene 8725 |
1 |
0.99 |
| chr5_90899182_90899382 | 0.31 |
Cxcl2 |
chemokine (C-X-C motif) ligand 2 |
4589 |
0.13 |
| chr17_26702508_26702992 | 0.30 |
Crebrf |
CREB3 regulatory factor |
12900 |
0.14 |
| chr3_60111256_60111413 | 0.30 |
Gm24382 |
predicted gene, 24382 |
16413 |
0.19 |
| chr10_4736776_4736976 | 0.30 |
Esr1 |
estrogen receptor 1 (alpha) |
24225 |
0.25 |
| chr2_4999219_4999388 | 0.30 |
Mcm10 |
minichromosome maintenance 10 replication initiation factor |
2255 |
0.21 |
| chr1_3515265_3515420 | 0.30 |
Gm37329 |
predicted gene, 37329 |
835 |
0.6 |
| chr2_147905769_147905939 | 0.29 |
Gm25516 |
predicted gene, 25516 |
30976 |
0.2 |
| chr3_118606217_118606485 | 0.29 |
Dpyd |
dihydropyrimidine dehydrogenase |
44165 |
0.15 |
| chr18_66004765_66005080 | 0.28 |
Lman1 |
lectin, mannose-binding, 1 |
2283 |
0.22 |
| chr6_24610468_24610637 | 0.28 |
Lmod2 |
leiomodin 2 (cardiac) |
12790 |
0.14 |
| chr4_94555776_94555981 | 0.28 |
Caap1 |
caspase activity and apoptosis inhibitor 1 |
888 |
0.53 |
| chr8_20817807_20818117 | 0.28 |
Gm20946 |
predicted gene, 20946 |
10391 |
0.15 |
| chr2_160848002_160848255 | 0.28 |
Gm11447 |
predicted gene 11447 |
2515 |
0.22 |
| chr4_108096711_108097042 | 0.27 |
Podn |
podocan |
431 |
0.78 |
| chr19_12684439_12684625 | 0.27 |
Gm49772 |
predicted gene, 49772 |
2761 |
0.13 |
| chr3_116302483_116302634 | 0.27 |
Gm29151 |
predicted gene 29151 |
47545 |
0.11 |
| chr2_11427432_11427583 | 0.27 |
Gm13296 |
predicted gene 13296 |
7283 |
0.12 |
| chr4_97900129_97900314 | 0.27 |
Nfia |
nuclear factor I/A |
10812 |
0.29 |
| chr3_152478706_152478877 | 0.27 |
Ak5 |
adenylate kinase 5 |
276 |
0.91 |
| chrY_90810953_90811113 | 0.26 |
Gm47283 |
predicted gene, 47283 |
20582 |
0.16 |
| chr4_136145338_136145489 | 0.26 |
Id3 |
inhibitor of DNA binding 3 |
439 |
0.76 |
| chr2_152830017_152830168 | 0.26 |
Bcl2l1 |
BCL2-like 1 |
218 |
0.9 |
| chr17_35815959_35816154 | 0.26 |
Ier3 |
immediate early response 3 |
5628 |
0.08 |
| chr6_149010896_149011065 | 0.26 |
Dennd5b |
DENN/MADD domain containing 5B |
6208 |
0.11 |
| chr9_94017367_94017557 | 0.25 |
Gm5369 |
predicted gene 5369 |
122000 |
0.06 |
| chr11_111998446_111998808 | 0.25 |
Gm11679 |
predicted gene 11679 |
44951 |
0.19 |
| chr1_15616194_15616345 | 0.25 |
Gm37138 |
predicted gene, 37138 |
60020 |
0.12 |
| chr6_137754996_137755471 | 0.25 |
Dera |
deoxyribose-phosphate aldolase (putative) |
618 |
0.77 |
| chr2_17326719_17326870 | 0.25 |
Nebl |
nebulette |
34292 |
0.22 |
| chr5_96304997_96305161 | 0.24 |
Fras1 |
Fraser extracellular matrix complex subunit 1 |
68876 |
0.11 |
| chr5_25496952_25497103 | 0.24 |
Kmt2c |
lysine (K)-specific methyltransferase 2C |
1675 |
0.26 |
| chr9_49485368_49485519 | 0.24 |
Ttc12 |
tetratricopeptide repeat domain 12 |
772 |
0.68 |
| chr1_162815471_162815637 | 0.24 |
Fmo4 |
flavin containing monooxygenase 4 |
1582 |
0.37 |
| chr16_37874612_37874787 | 0.24 |
Lrrc58 |
leucine rich repeat containing 58 |
6310 |
0.14 |
| chr2_164823177_164823502 | 0.24 |
Zswim1 |
zinc finger SWIM-type containing 1 |
326 |
0.72 |
| chrX_170020041_170020212 | 0.24 |
Erdr1 |
erythroid differentiation regulator 1 |
9382 |
0.18 |
| chr5_124424178_124424509 | 0.24 |
Gm37415 |
predicted gene, 37415 |
354 |
0.67 |
| chr10_4618857_4619069 | 0.24 |
Esr1 |
estrogen receptor 1 (alpha) |
6942 |
0.24 |
| chr6_144170659_144171019 | 0.23 |
Sox5 |
SRY (sex determining region Y)-box 5 |
33218 |
0.23 |
| chr9_96952383_96952534 | 0.23 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
19517 |
0.15 |
| chr17_53650070_53650394 | 0.23 |
Kat2b |
K(lysine) acetyltransferase 2B |
5535 |
0.15 |
| chr16_43247311_43247635 | 0.23 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
189 |
0.95 |
| chr10_89450844_89451017 | 0.23 |
Gas2l3 |
growth arrest-specific 2 like 3 |
6963 |
0.25 |
| chr1_131287169_131287361 | 0.22 |
Ikbke |
inhibitor of kappaB kinase epsilon |
7659 |
0.12 |
| chr4_115411935_115412583 | 0.22 |
Cyp4a12b |
cytochrome P450, family 4, subfamily a, polypeptide 12B |
635 |
0.62 |
| chr2_164447965_164448116 | 0.22 |
Sdc4 |
syndecan 4 |
4153 |
0.1 |
| chr18_33352007_33352191 | 0.22 |
Gm5503 |
predicted gene 5503 |
32856 |
0.21 |
| chr4_55352435_55352713 | 0.22 |
Rad23b |
RAD23 homolog B, nucleotide excision repair protein |
2484 |
0.24 |
| chr6_149147053_149147326 | 0.22 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
5543 |
0.14 |
| chr17_80772516_80772751 | 0.22 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
44148 |
0.15 |
| chr8_20289022_20289260 | 0.21 |
6820431F20Rik |
RIKEN cDNA 6820431F20 gene |
5875 |
0.25 |
| chr3_105773852_105774003 | 0.21 |
Gm43329 |
predicted gene 43329 |
3730 |
0.13 |
| chr4_136246535_136246839 | 0.21 |
Tcea3 |
transcription elongation factor A (SII), 3 |
1042 |
0.46 |
| chr5_51537712_51537892 | 0.21 |
Ppargc1a |
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
16069 |
0.16 |
| chr9_95540268_95540439 | 0.21 |
Gm32281 |
predicted gene, 32281 |
14144 |
0.13 |
| chr9_62087393_62087544 | 0.21 |
Glce |
glucuronyl C5-epimerase |
16628 |
0.19 |
| chr4_57122143_57122466 | 0.21 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
2244 |
0.35 |
| chr5_87457817_87457968 | 0.21 |
Ugt2a2 |
UDP glucuronosyltransferase 2 family, polypeptide A2 |
24366 |
0.1 |
| chr16_58662034_58662449 | 0.21 |
Gm49701 |
predicted gene, 49701 |
6997 |
0.13 |
| chr1_71807937_71808302 | 0.21 |
Gm37217 |
predicted gene, 37217 |
38491 |
0.15 |
| chr5_90519183_90519413 | 0.20 |
Afm |
afamin |
349 |
0.81 |
| chr12_32768098_32768327 | 0.20 |
Nampt |
nicotinamide phosphoribosyltransferase |
51333 |
0.12 |
| chr6_24964522_24964682 | 0.20 |
Tmem229a |
transmembrane protein 229A |
8305 |
0.22 |
| chr3_97653684_97653893 | 0.20 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
4405 |
0.15 |
| chr8_127463314_127463465 | 0.20 |
Pard3 |
par-3 family cell polarity regulator |
15643 |
0.28 |
| chr10_115386783_115386953 | 0.20 |
Zfc3h1 |
zinc finger, C3H1-type containing |
1909 |
0.24 |
| chr1_67133525_67134193 | 0.20 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
10833 |
0.24 |
| chr2_77173287_77173438 | 0.20 |
Ccdc141 |
coiled-coil domain containing 141 |
2726 |
0.29 |
| chr2_58782728_58782879 | 0.20 |
Upp2 |
uridine phosphorylase 2 |
17478 |
0.19 |
| chr3_144255924_144256075 | 0.20 |
Gm43446 |
predicted gene 43446 |
27812 |
0.16 |
| chr18_60243095_60243283 | 0.20 |
Gm5970 |
predicted gene 5970 |
22346 |
0.13 |
| chr9_66600725_66600966 | 0.19 |
Usp3 |
ubiquitin specific peptidase 3 |
7703 |
0.2 |
| chr5_86942001_86942153 | 0.19 |
Gm24121 |
predicted gene, 24121 |
13069 |
0.09 |
| chr1_170608002_170608165 | 0.19 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
18222 |
0.17 |
| chr2_109684188_109684342 | 0.19 |
Bdnf |
brain derived neurotrophic factor |
7233 |
0.19 |
| chr2_81270788_81270939 | 0.19 |
Gm23900 |
predicted gene, 23900 |
272479 |
0.01 |
| chrY_90794490_90794856 | 0.19 |
Gm47283 |
predicted gene, 47283 |
4222 |
0.21 |
| chr19_34868908_34869317 | 0.19 |
Pank1 |
pantothenate kinase 1 |
8737 |
0.17 |
| chr9_67845546_67845728 | 0.19 |
Vps13c |
vacuolar protein sorting 13C |
5225 |
0.21 |
| chr4_105179161_105179312 | 0.19 |
Plpp3 |
phospholipid phosphatase 3 |
21889 |
0.24 |
| chr1_182959595_182959833 | 0.19 |
Tlr5 |
toll-like receptor 5 |
4886 |
0.22 |
| chr6_140199181_140199332 | 0.19 |
Gm24174 |
predicted gene, 24174 |
32611 |
0.16 |
| chr11_119048222_119048411 | 0.19 |
Cbx8 |
chromobox 8 |
7347 |
0.16 |
| chr11_30773137_30773288 | 0.19 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
891 |
0.53 |
| chr6_72431165_72431316 | 0.18 |
Mat2a |
methionine adenosyltransferase II, alpha |
3751 |
0.13 |
| chrX_170022175_170022352 | 0.18 |
Erdr1 |
erythroid differentiation regulator 1 |
11519 |
0.18 |
| chr6_143513821_143514027 | 0.18 |
4930579D09Rik |
RIKEN cDNA 4930579D09 gene |
21584 |
0.23 |
| chr9_74326904_74327074 | 0.18 |
Gm24141 |
predicted gene, 24141 |
35621 |
0.16 |
| chr18_35499140_35499327 | 0.18 |
Sil1 |
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
304 |
0.64 |
| chr5_137778433_137778610 | 0.18 |
Ppp1r35 |
protein phosphatase 1, regulatory subunit 35 |
328 |
0.76 |
| chr1_65156847_65157132 | 0.18 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
18575 |
0.11 |
| chr17_70661451_70661755 | 0.17 |
5031415H12Rik |
RIKEN cDNA 5031415H12 gene |
93979 |
0.07 |
| chr1_131990500_131990816 | 0.17 |
Slc45a3 |
solute carrier family 45, member 3 |
11694 |
0.12 |
| chr1_21299385_21299536 | 0.17 |
Gm4956 |
predicted gene 4956 |
1148 |
0.32 |
| chr8_77100242_77100465 | 0.17 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
27660 |
0.19 |
| chr5_90696399_90696550 | 0.17 |
Gm43090 |
predicted gene 43090 |
54051 |
0.08 |
| chr15_103307817_103307990 | 0.17 |
Gpr84 |
G protein-coupled receptor 84 |
2709 |
0.14 |
| chr7_119960819_119960970 | 0.17 |
Dnah3 |
dynein, axonemal, heavy chain 3 |
6948 |
0.16 |
| chr3_51917272_51917733 | 0.17 |
Gm10728 |
predicted gene 10728 |
8686 |
0.13 |
| chr3_106496243_106496679 | 0.17 |
Dennd2d |
DENN/MADD domain containing 2D |
3286 |
0.17 |
| chr9_82938706_82938857 | 0.17 |
Phip |
pleckstrin homology domain interacting protein |
5749 |
0.26 |
| chr17_25447727_25447880 | 0.17 |
Tekt4 |
tektin 4 |
6812 |
0.12 |
| chr2_23090654_23090815 | 0.16 |
Acbd5 |
acyl-Coenzyme A binding domain containing 5 |
3467 |
0.23 |
| chr14_76800394_76800719 | 0.16 |
Gm30246 |
predicted gene, 30246 |
19729 |
0.17 |
| chr8_89207204_89207384 | 0.16 |
Gm5356 |
predicted pseudogene 5356 |
19734 |
0.24 |
| chr10_24362809_24362960 | 0.16 |
Gm15271 |
predicted gene 15271 |
84606 |
0.08 |
| chr3_37687682_37687873 | 0.16 |
Gm43414 |
predicted gene 43414 |
20244 |
0.1 |
| chr16_44149582_44149733 | 0.16 |
Naa50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
9765 |
0.17 |
| chr6_13676526_13676747 | 0.16 |
Bmt2 |
base methyltransferase of 25S rRNA 2 |
1302 |
0.55 |
| chr5_146330559_146330717 | 0.16 |
Cdk8 |
cyclin-dependent kinase 8 |
34273 |
0.13 |
| chr16_6901477_6901805 | 0.16 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
92419 |
0.1 |
| chr8_68008523_68008836 | 0.16 |
Gm22018 |
predicted gene, 22018 |
1661 |
0.43 |
| chr13_23849104_23849255 | 0.16 |
Slc17a3 |
solute carrier family 17 (sodium phosphate), member 3 |
9589 |
0.08 |
| chr9_106727325_106727482 | 0.16 |
Rad54l2 |
RAD54 like 2 (S. cerevisiae) |
4532 |
0.19 |
| chrX_109011797_109011968 | 0.16 |
Hmgn5 |
high-mobility group nucleosome binding domain 5 |
1522 |
0.29 |
| chr9_22467234_22467805 | 0.16 |
Rp9 |
retinitis pigmentosa 9 (human) |
651 |
0.58 |
| chr9_108797332_108797857 | 0.16 |
Ip6k2 |
inositol hexaphosphate kinase 2 |
593 |
0.53 |
| chr8_44937447_44937668 | 0.16 |
Fat1 |
FAT atypical cadherin 1 |
2110 |
0.3 |
| chr15_77929755_77929928 | 0.15 |
Txn2 |
thioredoxin 2 |
835 |
0.54 |
| chr12_91731624_91731976 | 0.15 |
Ston2 |
stonin 2 |
12055 |
0.17 |
| chr15_27461169_27461365 | 0.15 |
Gm36899 |
predicted gene, 36899 |
2182 |
0.26 |
| chr3_121778871_121779078 | 0.15 |
4633401B06Rik |
RIKEN cDNA 4633401B06 gene |
9598 |
0.11 |
| chr8_119925715_119925866 | 0.15 |
Usp10 |
ubiquitin specific peptidase 10 |
11892 |
0.16 |
| chr11_74648732_74649171 | 0.15 |
Cluh |
clustered mitochondria (cluA/CLU1) homolog |
544 |
0.74 |
| chr5_142614394_142614571 | 0.15 |
Mmd2 |
monocyte to macrophage differentiation-associated 2 |
5682 |
0.16 |
| chr4_36131854_36132041 | 0.15 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
4516 |
0.35 |
| chrY_90796785_90796987 | 0.15 |
Gm47283 |
predicted gene, 47283 |
6435 |
0.19 |
| chr4_149989662_149989976 | 0.15 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
11446 |
0.15 |
| chr10_71177574_71177846 | 0.15 |
1700113B09Rik |
RIKEN cDNA 1700113B09 gene |
3326 |
0.15 |
| chr6_37705705_37706037 | 0.15 |
Ybx1-ps2 |
Y box protein 1, pseudogene 2 |
18676 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.5 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.3 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.1 | 0.2 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.3 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.1 | 0.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.3 | GO:0080032 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |