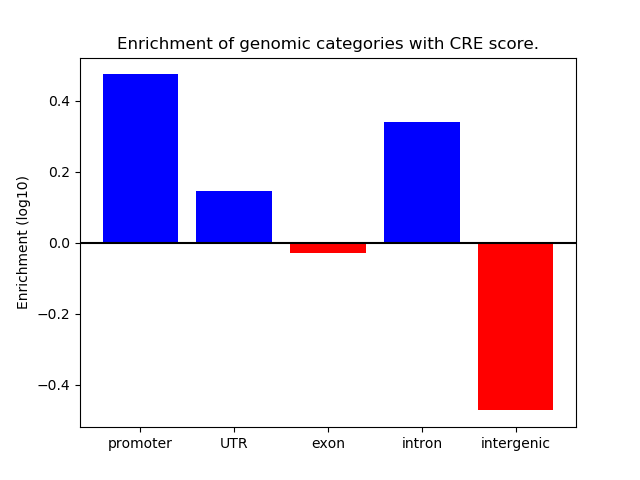
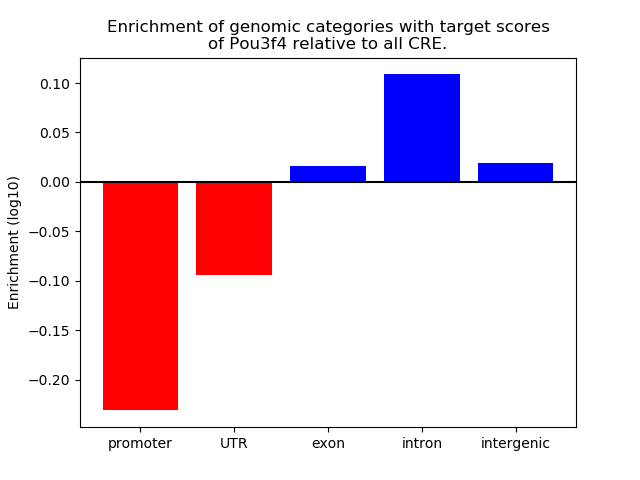
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
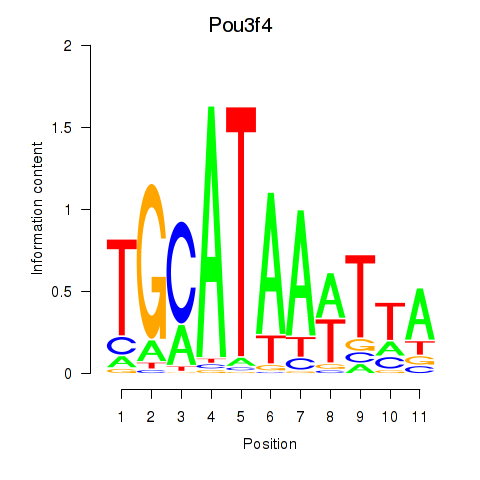
Results for Pou3f4
Z-value: 1.18
Transcription factors associated with Pou3f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f4
|
ENSMUSG00000056854.3 | POU domain, class 3, transcription factor 4 |
Activity of the Pou3f4 motif across conditions
Conditions sorted by the z-value of the Pou3f4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
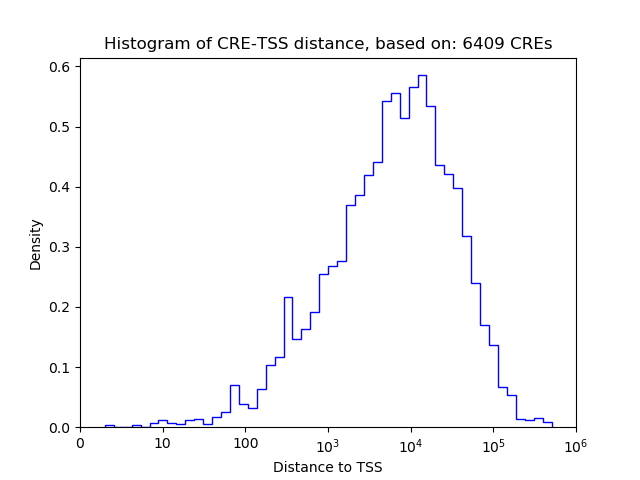
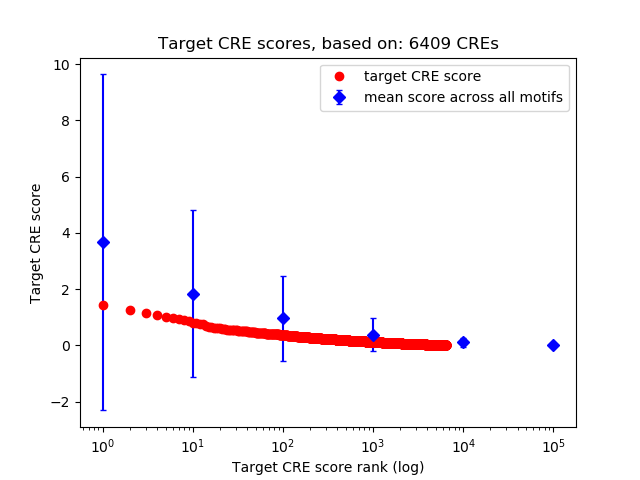
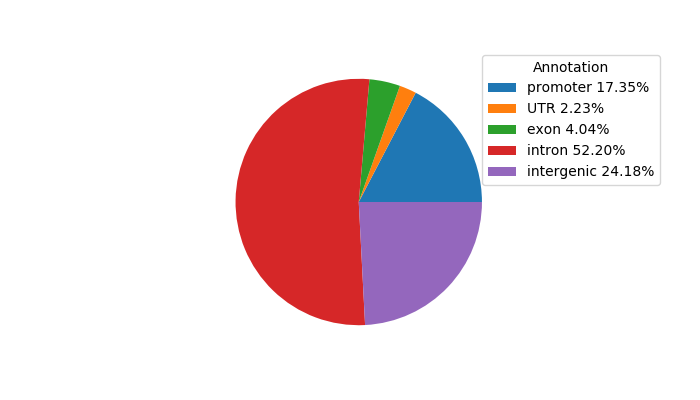
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_145775927_145776137 | 1.43 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
15284 |
0.2 |
| chr2_177500562_177500734 | 1.26 |
Gm14403 |
predicted gene 14403 |
2333 |
0.26 |
| chr11_11946176_11946345 | 1.16 |
Grb10 |
growth factor receptor bound protein 10 |
70 |
0.98 |
| chr19_40153747_40153898 | 1.08 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr19_40159590_40159741 | 1.02 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
27621 |
0.14 |
| chr8_105104839_105104994 | 0.95 |
Gm8804 |
predicted gene 8804 |
5708 |
0.11 |
| chr12_104343910_104344587 | 0.94 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
5762 |
0.12 |
| chr11_111269853_111270023 | 0.90 |
Gm11675 |
predicted gene 11675 |
6162 |
0.34 |
| chr2_73628966_73629117 | 0.87 |
Chn1 |
chimerin 1 |
3301 |
0.21 |
| chr11_5905326_5905477 | 0.80 |
Gck |
glucokinase |
3314 |
0.14 |
| chr10_24362809_24362960 | 0.78 |
Gm15271 |
predicted gene 15271 |
84606 |
0.08 |
| chr13_82200374_82200525 | 0.76 |
Gm48155 |
predicted gene, 48155 |
110692 |
0.07 |
| chr9_74893793_74894014 | 0.76 |
Onecut1 |
one cut domain, family member 1 |
27419 |
0.13 |
| chr18_38370754_38370932 | 0.70 |
Gm4949 |
predicted gene 4949 |
10305 |
0.12 |
| chr10_87880307_87880601 | 0.67 |
Igf1os |
insulin-like growth factor 1, opposite strand |
17073 |
0.18 |
| chr8_93183570_93183871 | 0.66 |
Gm45909 |
predicted gene 45909 |
7638 |
0.14 |
| chr7_35748679_35748830 | 0.62 |
Dpy19l3 |
dpy-19-like 3 (C. elegans) |
5601 |
0.2 |
| chr8_84761164_84761339 | 0.61 |
Nfix |
nuclear factor I/X |
12145 |
0.11 |
| chr4_108093158_108093452 | 0.61 |
Podn |
podocan |
3140 |
0.19 |
| chr12_57317079_57317230 | 0.61 |
Mipol1 |
mirror-image polydactyly 1 |
11071 |
0.22 |
| chr6_149147053_149147326 | 0.59 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
5543 |
0.14 |
| chr1_162987951_162988105 | 0.58 |
Fmo3 |
flavin containing monooxygenase 3 |
3500 |
0.2 |
| chr19_40165767_40165918 | 0.57 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
21444 |
0.14 |
| chr15_62744471_62744634 | 0.56 |
Gm22521 |
predicted gene, 22521 |
48649 |
0.17 |
| chr10_24362567_24362800 | 0.55 |
Gm15271 |
predicted gene 15271 |
84807 |
0.08 |
| chr12_32766682_32766844 | 0.55 |
Nampt |
nicotinamide phosphoribosyltransferase |
52782 |
0.12 |
| chr7_26817080_26817232 | 0.55 |
Cyp2g1 |
cytochrome P450, family 2, subfamily g, polypeptide 1 |
861 |
0.56 |
| chr13_112331091_112331250 | 0.54 |
Ankrd55 |
ankyrin repeat domain 55 |
12716 |
0.17 |
| chr12_86515829_86515996 | 0.54 |
Esrrb |
estrogen related receptor, beta |
11503 |
0.25 |
| chr5_107123329_107123511 | 0.54 |
Tgfbr3 |
transforming growth factor, beta receptor III |
9503 |
0.17 |
| chr8_93164567_93164980 | 0.53 |
Ces1d |
carboxylesterase 1D |
5202 |
0.15 |
| chr1_102517902_102518283 | 0.52 |
Gm20281 |
predicted gene, 20281 |
59170 |
0.14 |
| chr5_3492308_3492470 | 0.52 |
Gm42435 |
predicted gene 42435 |
7683 |
0.13 |
| chr4_135221446_135221961 | 0.52 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
51111 |
0.09 |
| chr11_16865620_16865819 | 0.52 |
Egfr |
epidermal growth factor receptor |
12431 |
0.2 |
| chr12_99858543_99858709 | 0.51 |
Efcab11 |
EF-hand calcium binding domain 11 |
24231 |
0.11 |
| chr5_125522690_125522841 | 0.50 |
Aacs |
acetoacetyl-CoA synthetase |
7522 |
0.16 |
| chr2_58777467_58777618 | 0.50 |
Upp2 |
uridine phosphorylase 2 |
12217 |
0.2 |
| chr8_93165222_93165456 | 0.50 |
Ces1d |
carboxylesterase 1D |
4636 |
0.15 |
| chr5_87570105_87570260 | 0.50 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
1155 |
0.34 |
| chr7_114356842_114356993 | 0.48 |
4933406I18Rik |
RIKEN cDNA 4933406I18 gene |
58104 |
0.11 |
| chr12_104083020_104083260 | 0.48 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr9_70446864_70447026 | 0.48 |
Rnf111 |
ring finger 111 |
6901 |
0.16 |
| chr12_45069728_45070323 | 0.47 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
4087 |
0.24 |
| chr10_127702413_127702583 | 0.47 |
Myo1a |
myosin IA |
2672 |
0.12 |
| chr2_73491256_73491419 | 0.47 |
Wipf1 |
WAS/WASL interacting protein family, member 1 |
4868 |
0.19 |
| chr19_53781327_53781513 | 0.47 |
Rbm20 |
RNA binding motif protein 20 |
11888 |
0.17 |
| chr19_26823641_26823967 | 0.46 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
103 |
0.97 |
| chr5_87589802_87589979 | 0.46 |
Sult1e1 |
sulfotransferase family 1E, member 1 |
1704 |
0.22 |
| chr19_44399803_44399962 | 0.45 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6808 |
0.15 |
| chr1_151124767_151124929 | 0.45 |
Gm8941 |
predicted gene 8941 |
11659 |
0.12 |
| chr1_58945674_58946262 | 0.45 |
Trak2 |
trafficking protein, kinesin binding 2 |
369 |
0.84 |
| chr9_106727325_106727482 | 0.45 |
Rad54l2 |
RAD54 like 2 (S. cerevisiae) |
4532 |
0.19 |
| chr2_72160606_72160802 | 0.45 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
11908 |
0.19 |
| chr3_41138122_41138276 | 0.45 |
Gm40038 |
predicted gene, 40038 |
34464 |
0.16 |
| chr16_90199102_90199289 | 0.44 |
Gm49704 |
predicted gene, 49704 |
1047 |
0.47 |
| chr6_114877506_114877696 | 0.44 |
Vgll4 |
vestigial like family member 4 |
2494 |
0.3 |
| chr16_18187141_18187312 | 0.44 |
Mir6366 |
microRNA 6366 |
22056 |
0.08 |
| chr2_58783629_58783834 | 0.44 |
Upp2 |
uridine phosphorylase 2 |
18406 |
0.19 |
| chr11_28696545_28697113 | 0.43 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
15265 |
0.17 |
| chr16_37902225_37902391 | 0.43 |
Gpr156 |
G protein-coupled receptor 156 |
14188 |
0.14 |
| chr1_151121601_151121803 | 0.42 |
Gm19087 |
predicted gene, 19087 |
13787 |
0.12 |
| chr10_76531927_76532170 | 0.42 |
Lss |
lanosterol synthase |
366 |
0.79 |
| chr1_21261382_21261702 | 0.42 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8021 |
0.11 |
| chr1_151720665_151721037 | 0.41 |
2810414N06Rik |
RIKEN cDNA 2810414N06 gene |
34167 |
0.14 |
| chr4_76468484_76468676 | 0.41 |
Gm42303 |
predicted gene, 42303 |
17349 |
0.21 |
| chr3_51185131_51185304 | 0.41 |
Noct |
nocturnin |
39230 |
0.12 |
| chr4_35916085_35916245 | 0.41 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
70961 |
0.13 |
| chr17_28428613_28428954 | 0.40 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr2_8124301_8124468 | 0.40 |
Gm13254 |
predicted gene 13254 |
23481 |
0.29 |
| chr4_15023934_15024314 | 0.40 |
Gm11844 |
predicted gene 11844 |
1542 |
0.5 |
| chr15_59056231_59056593 | 0.40 |
Mtss1 |
MTSS I-BAR domain containing 1 |
948 |
0.63 |
| chr4_3373115_3373279 | 0.40 |
Gm11784 |
predicted gene 11784 |
13649 |
0.2 |
| chr7_4814980_4815131 | 0.40 |
Ube2s |
ubiquitin-conjugating enzyme E2S |
2465 |
0.12 |
| chr9_74326904_74327074 | 0.40 |
Gm24141 |
predicted gene, 24141 |
35621 |
0.16 |
| chr1_21266470_21266781 | 0.40 |
Gm28836 |
predicted gene 28836 |
4968 |
0.12 |
| chr3_18137155_18137306 | 0.40 |
Gm23686 |
predicted gene, 23686 |
40395 |
0.15 |
| chr3_89896247_89896398 | 0.40 |
Gm42809 |
predicted gene 42809 |
16594 |
0.11 |
| chr6_85810872_85811030 | 0.40 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
1088 |
0.31 |
| chr1_164435834_164435997 | 0.39 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
7804 |
0.15 |
| chr5_150918259_150918412 | 0.39 |
Kl |
klotho |
34272 |
0.15 |
| chr4_35533897_35534048 | 0.39 |
Gm12365 |
predicted gene 12365 |
97758 |
0.08 |
| chr15_62533627_62533818 | 0.39 |
Gm41333 |
predicted gene, 41333 |
34253 |
0.23 |
| chr3_116302483_116302634 | 0.39 |
Gm29151 |
predicted gene 29151 |
47545 |
0.11 |
| chr16_37874612_37874787 | 0.39 |
Lrrc58 |
leucine rich repeat containing 58 |
6310 |
0.14 |
| chr2_58782728_58782879 | 0.39 |
Upp2 |
uridine phosphorylase 2 |
17478 |
0.19 |
| chr7_119957531_119957709 | 0.39 |
Dnah3 |
dynein, axonemal, heavy chain 3 |
5463 |
0.17 |
| chr11_111998446_111998808 | 0.39 |
Gm11679 |
predicted gene 11679 |
44951 |
0.19 |
| chr4_109801705_109801878 | 0.39 |
Faf1 |
Fas-associated factor 1 |
38994 |
0.16 |
| chr11_120804691_120805015 | 0.38 |
Fasn |
fatty acid synthase |
3752 |
0.1 |
| chr5_122017137_122017617 | 0.38 |
Gm3970 |
predicted gene 3970 |
7953 |
0.16 |
| chr1_67181891_67182275 | 0.38 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
59057 |
0.11 |
| chr19_12700970_12701121 | 0.38 |
Keg1 |
kidney expressed gene 1 |
5231 |
0.11 |
| chr15_3511597_3511812 | 0.38 |
Ghr |
growth hormone receptor |
40060 |
0.19 |
| chr13_62893171_62893343 | 0.38 |
Fbp1 |
fructose bisphosphatase 1 |
4975 |
0.16 |
| chr3_14832193_14832481 | 0.37 |
Car1 |
carbonic anhydrase 1 |
23969 |
0.15 |
| chr9_74328635_74328791 | 0.37 |
Gm24141 |
predicted gene, 24141 |
33897 |
0.17 |
| chr4_48261059_48261322 | 0.37 |
Erp44 |
endoplasmic reticulum protein 44 |
16716 |
0.18 |
| chr4_123990393_123990580 | 0.37 |
Gm12902 |
predicted gene 12902 |
64252 |
0.08 |
| chr3_141967234_141967427 | 0.37 |
Bmpr1b |
bone morphogenetic protein receptor, type 1B |
14934 |
0.29 |
| chr1_159269464_159269648 | 0.37 |
Cop1 |
COP1, E3 ubiquitin ligase |
2822 |
0.24 |
| chr19_40164076_40164449 | 0.37 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
23024 |
0.14 |
| chr19_17311642_17311793 | 0.37 |
Gcnt1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
23719 |
0.17 |
| chr5_142614394_142614571 | 0.36 |
Mmd2 |
monocyte to macrophage differentiation-associated 2 |
5682 |
0.16 |
| chr4_134837028_134837206 | 0.36 |
Maco1 |
macoilin 1 |
16032 |
0.17 |
| chr12_98524945_98525096 | 0.36 |
Gm40893 |
predicted gene, 40893 |
1176 |
0.44 |
| chr8_26078485_26078636 | 0.36 |
Hook3 |
hook microtubule tethering protein 3 |
817 |
0.46 |
| chr12_103937089_103937240 | 0.36 |
Gm17198 |
predicted gene 17198 |
10873 |
0.1 |
| chr10_4605236_4605428 | 0.36 |
Esr1 |
estrogen receptor 1 (alpha) |
6261 |
0.23 |
| chr8_80982761_80982912 | 0.35 |
Gm9725 |
predicted gene 9725 |
30740 |
0.13 |
| chr3_18148711_18148862 | 0.35 |
Gm23686 |
predicted gene, 23686 |
28839 |
0.19 |
| chr2_58773911_58774203 | 0.35 |
Upp2 |
uridine phosphorylase 2 |
8732 |
0.21 |
| chr4_100790971_100791127 | 0.35 |
Cachd1 |
cache domain containing 1 |
14374 |
0.28 |
| chr10_69208679_69208830 | 0.35 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
202 |
0.94 |
| chr2_17326719_17326870 | 0.34 |
Nebl |
nebulette |
34292 |
0.22 |
| chr5_96738098_96738279 | 0.34 |
Gm42604 |
predicted gene 42604 |
1176 |
0.5 |
| chr16_77416854_77417005 | 0.34 |
Gm38071 |
predicted gene, 38071 |
305 |
0.82 |
| chr3_148832133_148832284 | 0.34 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
1720 |
0.51 |
| chr14_57826803_57826983 | 0.34 |
Mrpl57 |
mitochondrial ribosomal protein L57 |
463 |
0.54 |
| chr4_53154081_53154232 | 0.33 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
5739 |
0.24 |
| chr12_32678889_32679054 | 0.33 |
Gm47937 |
predicted gene, 47937 |
5677 |
0.25 |
| chr3_58519897_58520081 | 0.33 |
Eif2a |
eukaryotic translation initiation factor 2A |
5832 |
0.16 |
| chr19_17761720_17761899 | 0.33 |
Gm17819 |
predicted gene, 17819 |
69046 |
0.1 |
| chr14_116240919_116241084 | 0.33 |
Gm20713 |
predicted gene 20713 |
369723 |
0.01 |
| chr18_76254679_76254860 | 0.33 |
Smad2 |
SMAD family member 2 |
6352 |
0.2 |
| chr11_16862337_16862489 | 0.33 |
Egfr |
epidermal growth factor receptor |
15737 |
0.19 |
| chr4_47366562_47366857 | 0.33 |
Tgfbr1 |
transforming growth factor, beta receptor I |
13099 |
0.22 |
| chr10_67192820_67193057 | 0.33 |
Jmjd1c |
jumonji domain containing 1C |
7185 |
0.22 |
| chr6_119739379_119739563 | 0.33 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
3973 |
0.32 |
| chr6_116079297_116079457 | 0.33 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
6221 |
0.18 |
| chr16_42908040_42908286 | 0.33 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
511 |
0.78 |
| chr11_120241116_120241542 | 0.33 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
4630 |
0.11 |
| chr10_78618331_78618509 | 0.32 |
Olfr1357 |
olfactory receptor 1357 |
346 |
0.74 |
| chr6_51695749_51695900 | 0.32 |
Gm38811 |
predicted gene, 38811 |
15257 |
0.23 |
| chr17_56573047_56573233 | 0.32 |
Safb2 |
scaffold attachment factor B2 |
642 |
0.57 |
| chr10_8642632_8642823 | 0.32 |
Gm24374 |
predicted gene, 24374 |
207 |
0.96 |
| chr2_72222188_72222339 | 0.32 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
10039 |
0.17 |
| chr4_134899949_134900100 | 0.32 |
Tmem50a |
transmembrane protein 50A |
14991 |
0.14 |
| chr6_114812242_114812393 | 0.32 |
Gm44331 |
predicted gene, 44331 |
22927 |
0.16 |
| chr5_28465358_28465587 | 0.32 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
1512 |
0.34 |
| chr6_85763479_85763641 | 0.32 |
Nat8f3 |
N-acetyltransferase 8 (GCN5-related) family member 3 |
1080 |
0.33 |
| chr16_37630277_37630428 | 0.32 |
Hgd |
homogentisate 1, 2-dioxygenase |
8503 |
0.16 |
| chr15_3496511_3496663 | 0.32 |
Ghr |
growth hormone receptor |
24943 |
0.24 |
| chr11_16696870_16697220 | 0.32 |
Gm25698 |
predicted gene, 25698 |
35666 |
0.13 |
| chr8_25256630_25256805 | 0.31 |
Tacc1 |
transforming, acidic coiled-coil containing protein 1 |
129 |
0.97 |
| chr2_122158054_122158205 | 0.31 |
Trim69 |
tripartite motif-containing 69 |
2571 |
0.17 |
| chr4_108096711_108097042 | 0.31 |
Podn |
podocan |
431 |
0.78 |
| chr16_24449611_24449762 | 0.31 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
1595 |
0.42 |
| chr2_122263270_122263421 | 0.31 |
Duox2 |
dual oxidase 2 |
22224 |
0.09 |
| chr5_114177314_114177465 | 0.31 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
1483 |
0.33 |
| chr2_109684188_109684342 | 0.31 |
Bdnf |
brain derived neurotrophic factor |
7233 |
0.19 |
| chr6_141854319_141854470 | 0.31 |
Slco1a4 |
solute carrier organic anion transporter family, member 1a4 |
712 |
0.71 |
| chr18_33435778_33435929 | 0.31 |
Nrep |
neuronal regeneration related protein |
27582 |
0.18 |
| chr11_72830025_72830176 | 0.31 |
Gm6733 |
predicted gene 6733 |
3361 |
0.19 |
| chr14_73382429_73382580 | 0.31 |
Itm2b |
integral membrane protein 2B |
2694 |
0.29 |
| chr4_90082681_90082858 | 0.30 |
Gm12629 |
predicted gene 12629 |
102758 |
0.08 |
| chr11_119048222_119048411 | 0.30 |
Cbx8 |
chromobox 8 |
7347 |
0.16 |
| chr18_61565305_61565485 | 0.30 |
Csnk1a1 |
casein kinase 1, alpha 1 |
9706 |
0.15 |
| chr6_15930296_15930597 | 0.30 |
Gm43990 |
predicted gene, 43990 |
94357 |
0.08 |
| chr6_144708083_144708633 | 0.30 |
Sox5os4 |
SRY (sex determining region Y)-box 5, opposite strand 4 |
12553 |
0.16 |
| chr16_57130407_57130558 | 0.30 |
Tomm70a |
translocase of outer mitochondrial membrane 70A |
7016 |
0.17 |
| chrX_151491088_151491256 | 0.30 |
Gm15148 |
predicted gene 15148 |
2097 |
0.3 |
| chr18_35458247_35458406 | 0.30 |
Gm50145 |
predicted gene, 50145 |
30778 |
0.1 |
| chr6_50091657_50091996 | 0.30 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
18415 |
0.22 |
| chr4_97100282_97100474 | 0.30 |
Gm27521 |
predicted gene, 27521 |
183358 |
0.03 |
| chr18_8933337_8933488 | 0.30 |
Gm37148 |
predicted gene, 37148 |
5177 |
0.27 |
| chr13_52819486_52819637 | 0.29 |
BB123696 |
expressed sequence BB123696 |
62356 |
0.13 |
| chr3_18107731_18107882 | 0.29 |
Gm23726 |
predicted gene, 23726 |
38480 |
0.15 |
| chr5_65999373_65999558 | 0.29 |
9130230L23Rik |
RIKEN cDNA 9130230L23 gene |
753 |
0.52 |
| chr5_100424142_100424293 | 0.29 |
5430416N02Rik |
RIKEN cDNA 5430416N02 gene |
5158 |
0.15 |
| chr10_14012990_14013158 | 0.29 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
46050 |
0.14 |
| chr3_105773852_105774003 | 0.29 |
Gm43329 |
predicted gene 43329 |
3730 |
0.13 |
| chr9_24502048_24502199 | 0.29 |
Dpy19l1 |
dpy-19-like 1 (C. elegans) |
1017 |
0.45 |
| chr1_105176971_105177328 | 0.29 |
Gm29012 |
predicted gene 29012 |
60695 |
0.12 |
| chr8_93165469_93165620 | 0.29 |
Ces1d |
carboxylesterase 1D |
4431 |
0.16 |
| chr10_50623725_50623913 | 0.29 |
Ascc3 |
activating signal cointegrator 1 complex subunit 3 |
18623 |
0.22 |
| chr16_56714341_56714503 | 0.29 |
Tfg |
Trk-fused gene |
1446 |
0.47 |
| chr5_107832772_107832923 | 0.29 |
Ube2d2b |
ubiquitin-conjugating enzyme E2D 2B |
2748 |
0.16 |
| chr9_74872523_74872674 | 0.29 |
Onecut1 |
one cut domain, family member 1 |
6114 |
0.16 |
| chr7_88307523_88307674 | 0.29 |
Gm44751 |
predicted gene 44751 |
3880 |
0.23 |
| chr8_22873332_22873513 | 0.29 |
Gm45555 |
predicted gene 45555 |
167 |
0.94 |
| chr1_69685147_69685314 | 0.29 |
Ikzf2 |
IKAROS family zinc finger 2 |
70 |
0.98 |
| chr1_56623913_56624064 | 0.28 |
Hsfy2 |
heat shock transcription factor, Y-linked 2 |
13447 |
0.28 |
| chr15_3397234_3397385 | 0.28 |
Ghr |
growth hormone receptor |
74335 |
0.1 |
| chr5_98992026_98992185 | 0.28 |
Prkg2 |
protein kinase, cGMP-dependent, type II |
25349 |
0.2 |
| chr17_71222209_71222384 | 0.28 |
Lpin2 |
lipin 2 |
9137 |
0.17 |
| chr7_46746742_46746969 | 0.28 |
Saa1 |
serum amyloid A 1 |
3875 |
0.1 |
| chr17_80022007_80022158 | 0.28 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr19_57872901_57873092 | 0.28 |
Mir5623 |
microRNA 5623 |
178171 |
0.03 |
| chr1_67133525_67134193 | 0.28 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
10833 |
0.24 |
| chr11_28694849_28695038 | 0.28 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
13379 |
0.17 |
| chr12_16875027_16875348 | 0.28 |
Rock2 |
Rho-associated coiled-coil containing protein kinase 2 |
19708 |
0.13 |
| chr14_117938790_117938941 | 0.28 |
Mir6239 |
microRNA 6239 |
14982 |
0.25 |
| chr5_87147107_87147418 | 0.28 |
Ugt2b5 |
UDP glucuronosyltransferase 2 family, polypeptide B5 |
6944 |
0.13 |
| chr11_28685236_28685405 | 0.28 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
3756 |
0.24 |
| chr3_31993839_31993990 | 0.28 |
Gm37834 |
predicted gene, 37834 |
39150 |
0.2 |
| chr3_149240098_149240249 | 0.28 |
Gm10287 |
predicted gene 10287 |
14428 |
0.2 |
| chr2_148027677_148027828 | 0.28 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
10518 |
0.17 |
| chr11_84185943_84186094 | 0.28 |
Gm37391 |
predicted gene, 37391 |
3575 |
0.21 |
| chr10_89472531_89472682 | 0.27 |
Gas2l3 |
growth arrest-specific 2 like 3 |
28639 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.3 | 0.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 1.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 0.7 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.4 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 1.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.5 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.1 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.3 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.1 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.3 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.3 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:1900094 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.0 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.0 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.4 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 1.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.9 | GO:0034892 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.4 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.0 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.0 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |