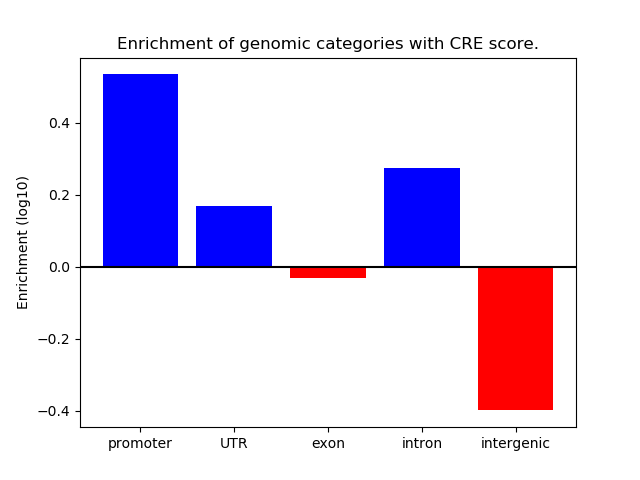
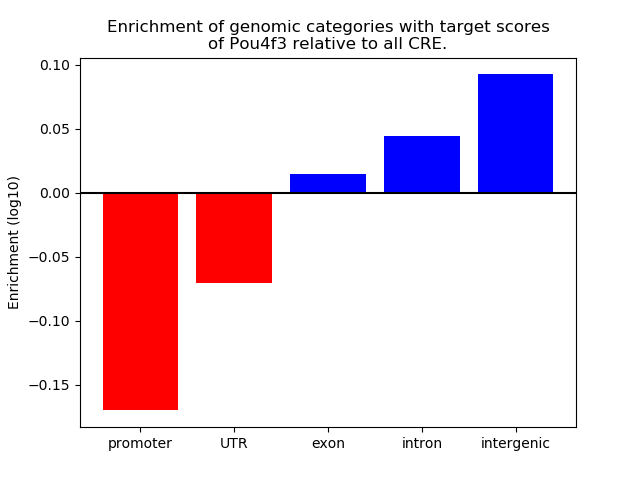
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
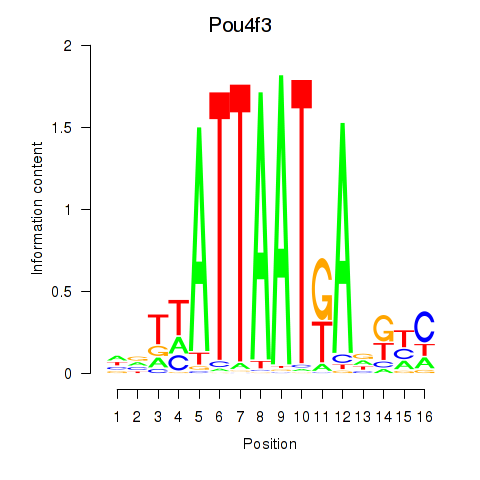
Results for Pou4f3
Z-value: 1.11
Transcription factors associated with Pou4f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f3
|
ENSMUSG00000024497.3 | POU domain, class 4, transcription factor 3 |
Activity of the Pou4f3 motif across conditions
Conditions sorted by the z-value of the Pou4f3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
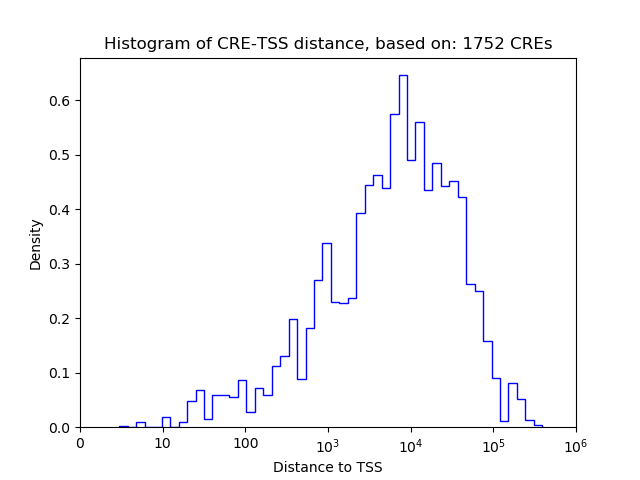
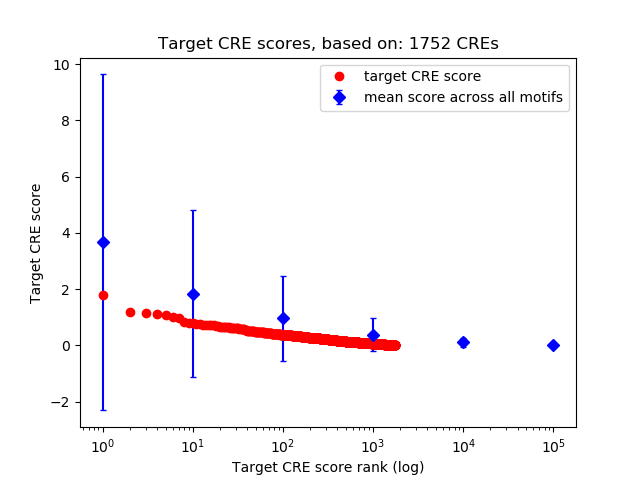
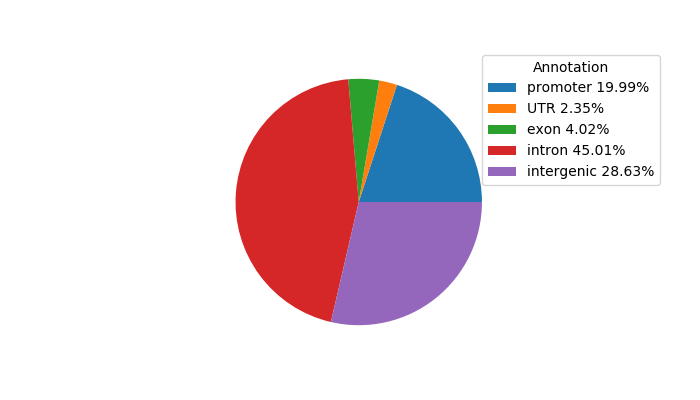
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_4616707_4616858 | 1.78 |
Lrfn4 |
leucine rich repeat and fibronectin type III domain containing 4 |
1115 |
0.34 |
| chr15_31279226_31279377 | 1.17 |
Gm49296 |
predicted gene, 49296 |
2788 |
0.19 |
| chr19_43971247_43971535 | 1.15 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr4_62508801_62509208 | 1.11 |
Hdhd3 |
haloacid dehalogenase-like hydrolase domain containing 3 |
5537 |
0.12 |
| chr19_46138051_46138212 | 1.09 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
842 |
0.48 |
| chr6_51392183_51392564 | 1.00 |
0610033M10Rik |
RIKEN cDNA 0610033M10 gene |
9660 |
0.17 |
| chr8_128113288_128113439 | 0.99 |
Mir21c |
microRNA 21c |
164862 |
0.04 |
| chr2_136986571_136986744 | 0.83 |
Slx4ip |
SLX4 interacting protein |
81114 |
0.08 |
| chr6_113828811_113828998 | 0.80 |
Gm44167 |
predicted gene, 44167 |
29220 |
0.13 |
| chr18_46336206_46336490 | 0.79 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
4081 |
0.17 |
| chr12_71430131_71430651 | 0.77 |
1700083H02Rik |
RIKEN cDNA 1700083H02 gene |
46183 |
0.12 |
| chr5_113041682_113041833 | 0.74 |
Gm22740 |
predicted gene, 22740 |
2387 |
0.2 |
| chr11_70713419_70713657 | 0.73 |
Mir6925 |
microRNA 6925 |
7548 |
0.08 |
| chr9_63201496_63201967 | 0.73 |
Skor1 |
SKI family transcriptional corepressor 1 |
52770 |
0.12 |
| chr8_126663289_126663717 | 0.72 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
69517 |
0.1 |
| chr9_7571293_7571444 | 0.72 |
Mmp27 |
matrix metallopeptidase 27 |
28 |
0.97 |
| chr5_145986007_145986356 | 0.70 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
5462 |
0.14 |
| chr10_75625748_75625899 | 0.69 |
Gm19688 |
predicted gene, 19688 |
6320 |
0.11 |
| chr6_71236030_71236205 | 0.68 |
Smyd1 |
SET and MYND domain containing 1 |
19243 |
0.1 |
| chr16_24394028_24394581 | 0.67 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr15_79690269_79690868 | 0.67 |
Gtpbp1 |
GTP binding protein 1 |
277 |
0.78 |
| chr16_93742880_93743193 | 0.65 |
Dop1b |
DOP1 leucine zipper like protein B |
12937 |
0.16 |
| chr10_68558838_68559075 | 0.65 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
17060 |
0.22 |
| chr11_109475236_109475399 | 0.64 |
Slc16a6 |
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
1719 |
0.22 |
| chr8_108101452_108101603 | 0.64 |
Zfhx3 |
zinc finger homeobox 3 |
106968 |
0.07 |
| chr4_45967836_45967998 | 0.63 |
Tdrd7 |
tudor domain containing 7 |
2583 |
0.3 |
| chr1_183979753_183980270 | 0.63 |
Dusp10 |
dual specificity phosphatase 10 |
33291 |
0.16 |
| chr3_94338287_94338715 | 0.63 |
Gm43743 |
predicted gene 43743 |
3169 |
0.1 |
| chr1_184622020_184622382 | 0.62 |
Gm37800 |
predicted gene, 37800 |
4230 |
0.18 |
| chr17_70746763_70747075 | 0.62 |
5031415H12Rik |
RIKEN cDNA 5031415H12 gene |
8663 |
0.19 |
| chr9_30613779_30614132 | 0.61 |
Gm47716 |
predicted gene, 47716 |
18632 |
0.24 |
| chr8_11149308_11149679 | 0.60 |
Gm44717 |
predicted gene 44717 |
247 |
0.91 |
| chr5_43157097_43157384 | 0.60 |
Gm42552 |
predicted gene 42552 |
8913 |
0.21 |
| chr19_44027305_44027461 | 0.60 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
1816 |
0.28 |
| chr8_113141217_113141373 | 0.58 |
Gm10280 |
predicted gene 10280 |
74033 |
0.12 |
| chr12_35284123_35284298 | 0.57 |
Gm44396 |
predicted gene, 44396 |
24681 |
0.22 |
| chr12_91929732_91929921 | 0.57 |
Rpl31-ps1 |
ribosomal protein L31, pseudogene 1 |
38553 |
0.11 |
| chr4_33229770_33229937 | 0.55 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
17722 |
0.14 |
| chr17_43299707_43299871 | 0.54 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
29460 |
0.2 |
| chr3_28736802_28736989 | 0.53 |
1700112D23Rik |
RIKEN cDNA 1700112D23 gene |
5345 |
0.16 |
| chr11_109500636_109500796 | 0.52 |
Gm22378 |
predicted gene, 22378 |
892 |
0.5 |
| chr3_98067419_98067570 | 0.52 |
Gm42819 |
predicted gene 42819 |
36807 |
0.13 |
| chr4_57525649_57525867 | 0.51 |
Pakap |
paralemmin A kinase anchor protein |
42401 |
0.16 |
| chr1_44130037_44130209 | 0.51 |
Bivm |
basic, immunoglobulin-like variable motif containing |
10144 |
0.12 |
| chr17_84037207_84037358 | 0.50 |
Gm49967 |
predicted gene, 49967 |
392 |
0.78 |
| chr4_137481328_137481504 | 0.50 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
12613 |
0.13 |
| chr4_60236200_60236578 | 0.49 |
Mup-ps3 |
major urinary protein, pseudogene 3 |
956 |
0.56 |
| chr10_43245095_43245284 | 0.49 |
Pdss2 |
prenyl (solanesyl) diphosphate synthase, subunit 2 |
23453 |
0.16 |
| chr11_121616042_121616235 | 0.49 |
Tbcd |
tubulin-specific chaperone d |
14433 |
0.2 |
| chr10_68269443_68270351 | 0.49 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
8824 |
0.23 |
| chr14_50991070_50991230 | 0.49 |
Gm49038 |
predicted gene, 49038 |
5925 |
0.08 |
| chr4_21739391_21739542 | 0.49 |
Ccnc |
cyclin C |
11699 |
0.16 |
| chr4_61317592_61317969 | 0.48 |
Mup-ps11 |
major urinary protein, pseudogene 11 |
950 |
0.58 |
| chr14_8331646_8331805 | 0.47 |
Fam107a |
family with sequence similarity 107, member A |
13702 |
0.18 |
| chr19_43767883_43768034 | 0.46 |
Cutc |
cutC copper transporter |
3085 |
0.18 |
| chr13_24950116_24950267 | 0.46 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
5663 |
0.14 |
| chr10_21665548_21665708 | 0.46 |
Gm5420 |
predicted gene 5420 |
20783 |
0.2 |
| chr15_81897988_81898152 | 0.46 |
Aco2 |
aconitase 2, mitochondrial |
2764 |
0.13 |
| chr4_60436201_60436591 | 0.46 |
Mup-ps4 |
major urinary protein, pseudogene 4 |
966 |
0.47 |
| chr7_140127944_140128095 | 0.46 |
Paox |
polyamine oxidase (exo-N4-amino) |
2311 |
0.14 |
| chr1_187214930_187215194 | 0.46 |
Spata17 |
spermatogenesis associated 17 |
359 |
0.55 |
| chr3_80042843_80043016 | 0.45 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
11963 |
0.22 |
| chr12_106479388_106479572 | 0.45 |
Gm3191 |
predicted gene 3191 |
20330 |
0.16 |
| chr5_49344702_49344867 | 0.45 |
Kcnip4 |
Kv channel interacting protein 4 |
59125 |
0.1 |
| chr5_138080510_138080792 | 0.44 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr1_88177823_88178019 | 0.44 |
AC087801.3 |
UDP glycosyltransferase 1 family (Ugt1) pseudogene |
380 |
0.64 |
| chr10_115337191_115337351 | 0.44 |
Tmem19 |
transmembrane protein 19 |
11793 |
0.14 |
| chr1_21228840_21229005 | 0.44 |
Gm38224 |
predicted gene, 38224 |
5800 |
0.13 |
| chr3_104220440_104221564 | 0.44 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
628 |
0.6 |
| chr16_4395267_4395559 | 0.43 |
Adcy9 |
adenylate cyclase 9 |
24174 |
0.18 |
| chr19_61057720_61057890 | 0.43 |
Gm22520 |
predicted gene, 22520 |
44260 |
0.12 |
| chr15_83454522_83454702 | 0.43 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
9940 |
0.15 |
| chr16_22919975_22920126 | 0.43 |
Fetub |
fetuin beta |
187 |
0.91 |
| chr10_21459651_21460332 | 0.42 |
Gm48386 |
predicted gene, 48386 |
14355 |
0.15 |
| chr17_30618531_30618854 | 0.42 |
Dnah8 |
dynein, axonemal, heavy chain 8 |
5662 |
0.11 |
| chr11_111605518_111605669 | 0.42 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr13_95777366_95777529 | 0.42 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
13050 |
0.19 |
| chr3_83155696_83155887 | 0.41 |
Gm10710 |
predicted gene 10710 |
26545 |
0.17 |
| chr6_124666813_124667013 | 0.41 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
3698 |
0.1 |
| chr16_38945803_38945963 | 0.41 |
Gm22500 |
predicted gene, 22500 |
18674 |
0.2 |
| chr1_131465972_131466299 | 0.41 |
Gm29487 |
predicted gene 29487 |
18440 |
0.14 |
| chr1_172514217_172514368 | 0.41 |
Gm37125 |
predicted gene, 37125 |
1043 |
0.35 |
| chr4_142088876_142089143 | 0.40 |
Tmem51os1 |
Tmem51 opposite strand 1 |
4153 |
0.16 |
| chr15_95897464_95897657 | 0.40 |
Gm25070 |
predicted gene, 25070 |
17743 |
0.17 |
| chr14_51100561_51100914 | 0.40 |
Rnase4 |
ribonuclease, RNase A family 4 |
4378 |
0.09 |
| chr10_94080659_94080847 | 0.40 |
Fgd6 |
FYVE, RhoGEF and PH domain containing 6 |
8211 |
0.13 |
| chr7_51887680_51888069 | 0.39 |
Gas2 |
growth arrest specific 2 |
44 |
0.97 |
| chr11_7519967_7520118 | 0.39 |
Gm11986 |
predicted gene 11986 |
229668 |
0.02 |
| chr1_77965517_77965856 | 0.39 |
Gm28387 |
predicted gene 28387 |
6690 |
0.29 |
| chr2_119566293_119566470 | 0.39 |
Chp1 |
calcineurin-like EF hand protein 1 |
224 |
0.89 |
| chr1_179352076_179352287 | 0.39 |
Smyd3 |
SET and MYND domain containing 3 |
21922 |
0.26 |
| chr1_21131249_21131442 | 0.39 |
Gm2693 |
predicted gene 2693 |
47643 |
0.1 |
| chr4_116720481_116720689 | 0.39 |
Tesk2 |
testis-specific kinase 2 |
363 |
0.78 |
| chr8_34506104_34506386 | 0.38 |
Gm8268 |
predicted gene 8268 |
962 |
0.54 |
| chr5_145464782_145464950 | 0.38 |
Cyp3a16 |
cytochrome P450, family 3, subfamily a, polypeptide 16 |
4857 |
0.19 |
| chr4_81841873_81842024 | 0.38 |
n-R5s187 |
nuclear encoded rRNA 5S 187 |
76565 |
0.1 |
| chr2_43548853_43549055 | 0.38 |
Kynu |
kynureninase |
6375 |
0.28 |
| chr11_86951820_86952000 | 0.38 |
Ypel2 |
yippee like 2 |
20114 |
0.17 |
| chr17_29381584_29381926 | 0.38 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
5254 |
0.14 |
| chr9_103363870_103364326 | 0.38 |
Cdv3 |
carnitine deficiency-associated gene expressed in ventricle 3 |
799 |
0.56 |
| chr9_43104579_43104740 | 0.37 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
827 |
0.61 |
| chr12_99418906_99419292 | 0.37 |
Foxn3 |
forkhead box N3 |
6494 |
0.18 |
| chr10_20046183_20046505 | 0.37 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
53181 |
0.13 |
| chr12_71899715_71899878 | 0.37 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
10066 |
0.23 |
| chr19_43768047_43768256 | 0.37 |
Cutc |
cutC copper transporter |
3278 |
0.18 |
| chr1_127011786_127011937 | 0.37 |
Gm5261 |
predicted gene 5261 |
38835 |
0.2 |
| chr13_74769953_74770127 | 0.37 |
Cast |
calpastatin |
550 |
0.67 |
| chr6_54849831_54850051 | 0.36 |
Znrf2 |
zinc and ring finger 2 |
32493 |
0.16 |
| chr4_62088107_62088281 | 0.36 |
Gm12909 |
predicted gene 12909 |
94 |
0.92 |
| chr1_133251724_133252043 | 0.36 |
Gm19461 |
predicted gene, 19461 |
2409 |
0.23 |
| chr19_38127249_38127411 | 0.36 |
Rbp4 |
retinol binding protein 4, plasma |
2049 |
0.25 |
| chr15_67062759_67062944 | 0.36 |
Gm31342 |
predicted gene, 31342 |
22793 |
0.2 |
| chr17_12924054_12924394 | 0.36 |
Snora20 |
small nucleolar RNA, H/ACA box 20 |
1434 |
0.19 |
| chr16_31986976_31987127 | 0.36 |
Gm49733 |
predicted gene, 49733 |
72 |
0.81 |
| chr19_5840987_5841190 | 0.36 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
4171 |
0.09 |
| chr4_35101216_35101367 | 0.36 |
Ifnk |
interferon kappa |
50765 |
0.12 |
| chr5_130264636_130264798 | 0.35 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
5787 |
0.13 |
| chr3_155092466_155092737 | 0.35 |
Fpgt |
fucose-1-phosphate guanylyltransferase |
46 |
0.94 |
| chr6_35874026_35874908 | 0.35 |
Gm43442 |
predicted gene 43442 |
52244 |
0.17 |
| chr1_88764125_88764295 | 0.35 |
Platr5 |
pluripotency associated transcript 5 |
9256 |
0.2 |
| chr4_55516147_55516328 | 0.35 |
Gm12507 |
predicted gene 12507 |
7943 |
0.16 |
| chr1_178330385_178330898 | 0.35 |
Hnrnpu |
heterogeneous nuclear ribonucleoprotein U |
20 |
0.96 |
| chr7_138913719_138913877 | 0.35 |
Gm9358 |
predicted gene 9358 |
590 |
0.61 |
| chr4_60676733_60676914 | 0.35 |
Mup-ps7 |
major urinary protein, pseudogene 7 |
860 |
0.58 |
| chr7_121469578_121469762 | 0.35 |
Gm36736 |
predicted gene, 36736 |
76569 |
0.08 |
| chr6_121874198_121874349 | 0.35 |
Mug1 |
murinoglobulin 1 |
11304 |
0.19 |
| chr6_12465944_12466328 | 0.34 |
Thsd7a |
thrombospondin, type I, domain containing 7A |
46775 |
0.18 |
| chr4_63738674_63738850 | 0.34 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
6351 |
0.22 |
| chrX_75409783_75409942 | 0.34 |
Cmc4 |
C-x(9)-C motif containing 4 |
50 |
0.95 |
| chrX_71331504_71331669 | 0.34 |
Mtm1 |
X-linked myotubular myopathy gene 1 |
30217 |
0.17 |
| chr11_60786182_60786455 | 0.34 |
Shmt1 |
serine hydroxymethyltransferase 1 (soluble) |
6855 |
0.08 |
| chr5_87091073_87091224 | 0.34 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
9 |
0.96 |
| chr9_32874453_32874604 | 0.34 |
Gm37167 |
predicted gene, 37167 |
34606 |
0.16 |
| chr4_60303896_60304116 | 0.34 |
Mup-ps3 |
major urinary protein, pseudogene 3 |
66661 |
0.08 |
| chr9_69291273_69291476 | 0.33 |
Rora |
RAR-related orphan receptor alpha |
1692 |
0.45 |
| chr15_90344050_90344371 | 0.33 |
Gm36480 |
predicted gene, 36480 |
9496 |
0.28 |
| chr5_66979311_66979467 | 0.33 |
Limch1 |
LIM and calponin homology domains 1 |
2371 |
0.2 |
| chr9_115403471_115403629 | 0.33 |
Gm9487 |
predicted gene 9487 |
1599 |
0.28 |
| chr1_184290067_184290274 | 0.33 |
Gm37223 |
predicted gene, 37223 |
68159 |
0.11 |
| chr7_79985891_79986050 | 0.33 |
Gm24541 |
predicted gene, 24541 |
36783 |
0.08 |
| chr19_36735506_36735662 | 0.33 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
1069 |
0.56 |
| chr1_136229022_136229181 | 0.33 |
Inava |
innate immunity activator |
918 |
0.4 |
| chr13_98578566_98578736 | 0.33 |
Gm22698 |
predicted gene, 22698 |
18297 |
0.12 |
| chr19_34518935_34519125 | 0.33 |
Lipa |
lysosomal acid lipase A |
8050 |
0.13 |
| chr2_159732589_159732753 | 0.33 |
Gm11445 |
predicted gene 11445 |
45536 |
0.19 |
| chr6_73281111_73281418 | 0.33 |
Suclg1 |
succinate-CoA ligase, GDP-forming, alpha subunit |
20904 |
0.16 |
| chr11_100836746_100836921 | 0.32 |
Stat5b |
signal transducer and activator of transcription 5B |
13705 |
0.12 |
| chrX_161939930_161940295 | 0.32 |
Gm15202 |
predicted gene 15202 |
31889 |
0.21 |
| chr10_93078296_93078483 | 0.32 |
Cfap54 |
cilia and flagella associated protein 54 |
3207 |
0.26 |
| chr5_16470617_16470792 | 0.32 |
Gm8984 |
predicted gene 8984 |
38479 |
0.17 |
| chr4_84102833_84103037 | 0.32 |
Gm12416 |
predicted gene 12416 |
1960 |
0.38 |
| chr14_46545888_46546175 | 0.32 |
E130120K24Rik |
RIKEN cDNA E130120K24 gene |
10272 |
0.13 |
| chr10_31644816_31644968 | 0.32 |
Gm8793 |
predicted gene 8793 |
5734 |
0.23 |
| chr11_78418219_78418374 | 0.32 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
3921 |
0.12 |
| chr6_47819662_47819827 | 0.32 |
Rpl35a-ps7 |
ribosomal protein L35A, pseudogene 7 |
4112 |
0.12 |
| chr12_25127702_25127874 | 0.32 |
Gm17746 |
predicted gene, 17746 |
1386 |
0.38 |
| chr18_39827979_39828276 | 0.31 |
Pabpc2 |
poly(A) binding protein, cytoplasmic 2 |
54630 |
0.14 |
| chr1_84877752_84878205 | 0.31 |
Fbxo36 |
F-box protein 36 |
8827 |
0.16 |
| chr8_71405347_71405498 | 0.31 |
Ankle1 |
ankyrin repeat and LEM domain containing 1 |
588 |
0.54 |
| chr15_70531676_70531845 | 0.31 |
Gm18155 |
predicted gene, 18155 |
226293 |
0.02 |
| chr13_98684591_98684906 | 0.31 |
Tmem171 |
transmembrane protein 171 |
10020 |
0.14 |
| chr1_105199808_105199959 | 0.31 |
Gm29013 |
predicted gene 29013 |
44542 |
0.16 |
| chr4_150685463_150685649 | 0.31 |
Gm16079 |
predicted gene 16079 |
6764 |
0.21 |
| chr11_83852736_83852913 | 0.31 |
Hnf1b |
HNF1 homeobox B |
136 |
0.94 |
| chr9_71163225_71163399 | 0.31 |
Aqp9 |
aquaporin 9 |
23 |
0.9 |
| chr9_115915920_115916106 | 0.31 |
Gadl1 |
glutamate decarboxylase-like 1 |
6558 |
0.28 |
| chr3_116936111_116936286 | 0.30 |
Gm42892 |
predicted gene 42892 |
8132 |
0.15 |
| chr4_141718181_141718503 | 0.30 |
Ddi2 |
DNA-damage inducible protein 2 |
5077 |
0.14 |
| chr3_27431035_27431235 | 0.30 |
Ghsr |
growth hormone secretagogue receptor |
59784 |
0.12 |
| chr4_132946310_132946569 | 0.30 |
Gm24913 |
predicted gene, 24913 |
15122 |
0.13 |
| chr19_5829011_5829567 | 0.30 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
1277 |
0.22 |
| chr4_61697893_61698073 | 0.30 |
Mup-ps15 |
major urinary protein, pseudogene 15 |
858 |
0.56 |
| chr13_55613655_55613852 | 0.30 |
Gm15911 |
predicted gene 15911 |
8970 |
0.1 |
| chr1_138992310_138992466 | 0.30 |
Gm16305 |
predicted gene 16305 |
319 |
0.81 |
| chr9_78501386_78501557 | 0.30 |
Eef1a1 |
eukaryotic translation elongation factor 1 alpha 1 |
12320 |
0.11 |
| chr12_78248706_78249176 | 0.30 |
Gm18899 |
predicted gene, 18899 |
7494 |
0.15 |
| chr17_36032663_36032990 | 0.29 |
H2-T23 |
histocompatibility 2, T region locus 23 |
29 |
0.92 |
| chr3_81995449_81996057 | 0.29 |
Asic5 |
acid-sensing (proton-gated) ion channel family member 5 |
1169 |
0.45 |
| chr15_91013020_91013330 | 0.29 |
Kif21a |
kinesin family member 21A |
36643 |
0.14 |
| chr2_120170818_120170979 | 0.29 |
Ehd4 |
EH-domain containing 4 |
16292 |
0.15 |
| chr10_21467545_21467696 | 0.29 |
Gm48386 |
predicted gene, 48386 |
21984 |
0.14 |
| chr8_64965219_64965377 | 0.29 |
Tmem192 |
transmembrane protein 192 |
5755 |
0.12 |
| chr14_75179701_75179900 | 0.29 |
Lcp1 |
lymphocyte cytosolic protein 1 |
3592 |
0.19 |
| chr10_20076891_20077084 | 0.29 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
22538 |
0.2 |
| chr2_165921904_165922055 | 0.29 |
Gm11461 |
predicted gene 11461 |
3862 |
0.16 |
| chr4_144957116_144957269 | 0.29 |
Gm38074 |
predicted gene, 38074 |
1656 |
0.38 |
| chr12_98860402_98860781 | 0.29 |
A930040O22Rik |
RIKEN cDNA A930040O22 gene |
28778 |
0.12 |
| chr1_58149355_58149553 | 0.28 |
Gm24548 |
predicted gene, 24548 |
5634 |
0.2 |
| chr2_32727271_32727422 | 0.28 |
Sh2d3c |
SH2 domain containing 3C |
360 |
0.69 |
| chr4_62015707_62015880 | 0.28 |
Mup-ps20 |
major urinary protein, pseudogene 20 |
936 |
0.5 |
| chr11_98339208_98339359 | 0.28 |
Ppp1r1b |
protein phosphatase 1, regulatory inhibitor subunit 1B |
9121 |
0.09 |
| chr17_5012388_5012845 | 0.28 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
16187 |
0.23 |
| chr6_94226332_94226483 | 0.28 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
56618 |
0.14 |
| chr7_38187215_38187367 | 0.28 |
1600014C10Rik |
RIKEN cDNA 1600014C10 gene |
1020 |
0.43 |
| chr1_156056827_156057192 | 0.28 |
Tor1aip2 |
torsin A interacting protein 2 |
6204 |
0.17 |
| chr3_122939334_122939509 | 0.28 |
Usp53 |
ubiquitin specific peptidase 53 |
10206 |
0.12 |
| chr2_57182280_57182573 | 0.28 |
4930555B11Rik |
RIKEN cDNA 4930555B11 gene |
595 |
0.46 |
| chr2_126551230_126551540 | 0.28 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
1022 |
0.56 |
| chr18_34328570_34328733 | 0.28 |
Srp19 |
signal recognition particle 19 |
2196 |
0.29 |
| chr16_91379054_91379459 | 0.27 |
Ifnar2 |
interferon (alpha and beta) receptor 2 |
6357 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.3 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.3 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.2 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.2 | GO:0009813 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.4 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |