Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
Results for Pou6f2_Pou4f2
Z-value: 0.57
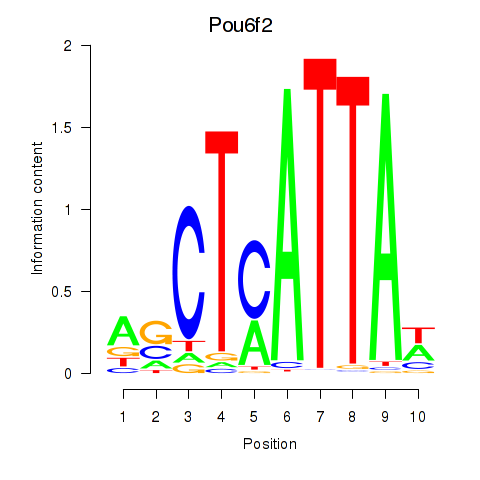
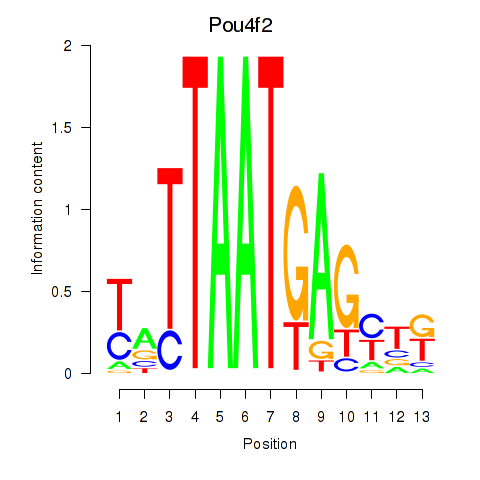
Transcription factors associated with Pou6f2_Pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou6f2
|
ENSMUSG00000009734.11 | POU domain, class 6, transcription factor 2 |
|
Pou4f2
|
ENSMUSG00000031688.3 | POU domain, class 4, transcription factor 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_18172902_18173081 | Pou6f2 | 451 | 0.852999 | 0.65 | 1.6e-01 | Click! |
| chr13_18173135_18173290 | Pou6f2 | 672 | 0.744534 | 0.54 | 2.7e-01 | Click! |
| chr13_18334141_18334292 | Pou6f2 | 47823 | 0.174335 | -0.25 | 6.3e-01 | Click! |
| chr13_18172418_18172600 | Pou6f2 | 26 | 0.981810 | 0.13 | 8.0e-01 | Click! |
| chr13_18174106_18174277 | Pou6f2 | 1651 | 0.424402 | 0.09 | 8.7e-01 | Click! |
Activity of the Pou6f2_Pou4f2 motif across conditions
Conditions sorted by the z-value of the Pou6f2_Pou4f2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
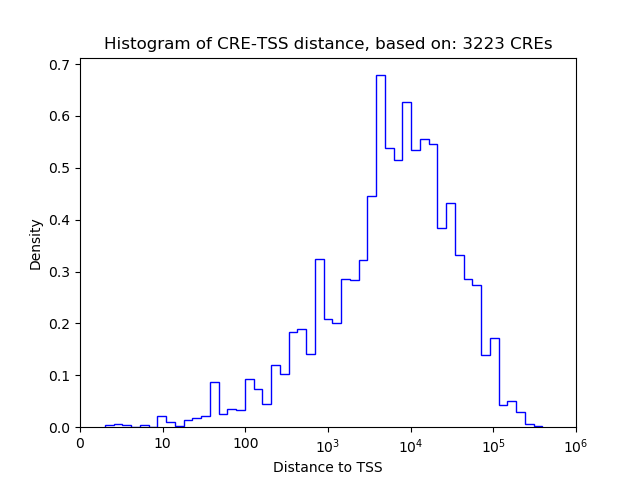
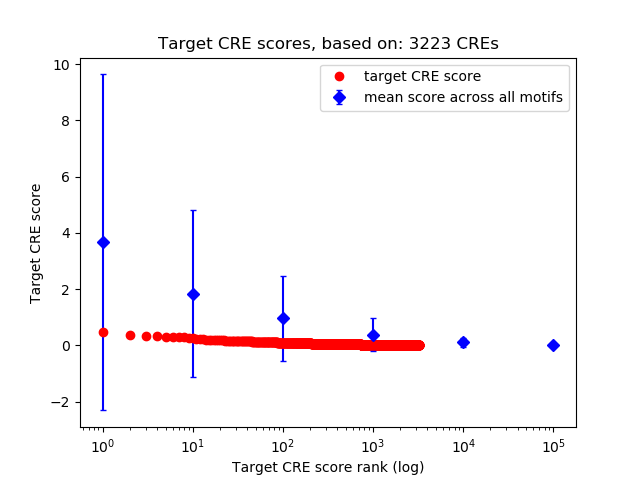
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_46138051_46138212 | 0.46 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
842 |
0.48 |
| chr5_49344702_49344867 | 0.35 |
Kcnip4 |
Kv channel interacting protein 4 |
59125 |
0.1 |
| chr16_31998270_31998421 | 0.34 |
Senp5 |
SUMO/sentrin specific peptidase 5 |
4818 |
0.09 |
| chr17_85028249_85028425 | 0.34 |
Slc3a1 |
solute carrier family 3, member 1 |
39 |
0.98 |
| chr2_62386509_62386700 | 0.31 |
Dpp4 |
dipeptidylpeptidase 4 |
8356 |
0.19 |
| chr18_20938375_20938560 | 0.30 |
Rnf125 |
ring finger protein 125 |
6158 |
0.22 |
| chr16_55810073_55810224 | 0.29 |
Nfkbiz |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
8367 |
0.18 |
| chr5_145997024_145997348 | 0.29 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
5543 |
0.14 |
| chr13_68712216_68712409 | 0.25 |
Gm26929 |
predicted gene, 26929 |
97768 |
0.06 |
| chr6_121878156_121878361 | 0.25 |
Mug1 |
murinoglobulin 1 |
7319 |
0.2 |
| chr9_33001470_33001748 | 0.23 |
Gm27166 |
predicted gene 27166 |
30182 |
0.17 |
| chr18_46336206_46336490 | 0.23 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
4081 |
0.17 |
| chr17_30632040_30632191 | 0.22 |
Dnah8 |
dynein, axonemal, heavy chain 8 |
3641 |
0.14 |
| chr11_112218841_112219019 | 0.20 |
Gm11680 |
predicted gene 11680 |
111492 |
0.07 |
| chr17_79451613_79451775 | 0.20 |
Cdc42ep3 |
CDC42 effector protein (Rho GTPase binding) 3 |
96603 |
0.07 |
| chr15_102078441_102078616 | 0.19 |
Eif4b |
eukaryotic translation initiation factor 4B |
4664 |
0.14 |
| chr9_121672421_121672607 | 0.19 |
Vipr1 |
vasoactive intestinal peptide receptor 1 |
3801 |
0.14 |
| chr5_126565776_126566015 | 0.18 |
Gm24839 |
predicted gene, 24839 |
51914 |
0.14 |
| chr15_22870784_22870938 | 0.18 |
Gm19840 |
predicted gene, 19840 |
64626 |
0.14 |
| chr9_65302674_65302855 | 0.18 |
Gm16218 |
predicted gene 16218 |
4845 |
0.11 |
| chr11_111605518_111605669 | 0.17 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr15_38661347_38661720 | 0.17 |
Atp6v1c1 |
ATPase, H+ transporting, lysosomal V1 subunit C1 |
400 |
0.79 |
| chr10_93518068_93518227 | 0.17 |
Hal |
histidine ammonia lyase |
17022 |
0.12 |
| chr19_43971247_43971535 | 0.16 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr10_95322653_95322993 | 0.16 |
Cradd |
CASP2 and RIPK1 domain containing adaptor with death domain |
142 |
0.94 |
| chr3_132863960_132864140 | 0.16 |
Gm29811 |
predicted gene, 29811 |
14546 |
0.14 |
| chr9_22945594_22945745 | 0.16 |
Gm27639 |
predicted gene, 27639 |
172834 |
0.03 |
| chr1_151158026_151158565 | 0.16 |
C730036E19Rik |
RIKEN cDNA C730036E19 gene |
20261 |
0.1 |
| chr10_20622440_20622618 | 0.16 |
Gm17230 |
predicted gene 17230 |
3106 |
0.32 |
| chr5_100860430_100860646 | 0.16 |
4930458D05Rik |
RIKEN cDNA 4930458D05 gene |
13371 |
0.1 |
| chr3_60495047_60495368 | 0.15 |
Mbnl1 |
muscleblind like splicing factor 1 |
5728 |
0.26 |
| chr1_184622020_184622382 | 0.15 |
Gm37800 |
predicted gene, 37800 |
4230 |
0.18 |
| chr6_54849831_54850051 | 0.15 |
Znrf2 |
zinc and ring finger 2 |
32493 |
0.16 |
| chr9_32752728_32752915 | 0.15 |
Gm27240 |
predicted gene 27240 |
41650 |
0.13 |
| chr12_71899715_71899878 | 0.15 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
10066 |
0.23 |
| chr18_46725311_46725732 | 0.15 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
2508 |
0.19 |
| chr16_26163678_26163843 | 0.15 |
P3h2 |
prolyl 3-hydroxylase 2 |
57976 |
0.15 |
| chr2_132093036_132093197 | 0.14 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
1513 |
0.37 |
| chr5_117165254_117165405 | 0.14 |
n-R5s174 |
nuclear encoded rRNA 5S 174 |
31226 |
0.11 |
| chr11_77215721_77215937 | 0.14 |
Ssh2 |
slingshot protein phosphatase 2 |
458 |
0.76 |
| chr12_3811591_3811763 | 0.14 |
Dnmt3a |
DNA methyltransferase 3A |
4517 |
0.2 |
| chr11_7201515_7201726 | 0.14 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
3838 |
0.2 |
| chr1_86206435_86206591 | 0.14 |
Armc9 |
armadillo repeat containing 9 |
4017 |
0.18 |
| chr12_117720585_117720755 | 0.14 |
Gm18955 |
predicted gene, 18955 |
3302 |
0.28 |
| chr7_19563093_19563255 | 0.14 |
Ppp1r37 |
protein phosphatase 1, regulatory subunit 37 |
98 |
0.93 |
| chr6_87428535_87428790 | 0.13 |
Bmp10 |
bone morphogenetic protein 10 |
332 |
0.85 |
| chr9_123530850_123531001 | 0.13 |
Sacm1l |
SAC1 suppressor of actin mutations 1-like (yeast) |
1043 |
0.5 |
| chr16_18715068_18715219 | 0.13 |
Rps2-ps7 |
ribosomal protein S2, pseudogene 7 |
32927 |
0.12 |
| chr15_77251530_77251759 | 0.13 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
6058 |
0.17 |
| chr8_122442495_122442674 | 0.13 |
Mvd |
mevalonate (diphospho) decarboxylase |
812 |
0.39 |
| chr5_9045029_9045180 | 0.13 |
Gm40264 |
predicted gene, 40264 |
9980 |
0.15 |
| chr6_113828811_113828998 | 0.13 |
Gm44167 |
predicted gene, 44167 |
29220 |
0.13 |
| chr6_112530748_112531068 | 0.13 |
Gm27944 |
predicted gene, 27944 |
17461 |
0.15 |
| chr3_57448063_57448221 | 0.13 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
22828 |
0.19 |
| chr15_67062759_67062944 | 0.13 |
Gm31342 |
predicted gene, 31342 |
22793 |
0.2 |
| chr16_4404498_4404756 | 0.12 |
Adcy9 |
adenylate cyclase 9 |
14960 |
0.2 |
| chr16_93742880_93743193 | 0.12 |
Dop1b |
DOP1 leucine zipper like protein B |
12937 |
0.16 |
| chr11_21024845_21024996 | 0.12 |
Peli1 |
pellino 1 |
66371 |
0.1 |
| chr3_79156332_79156483 | 0.12 |
Rapgef2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
10532 |
0.21 |
| chr11_104289393_104289544 | 0.12 |
Mapt |
microtubule-associated protein tau |
7087 |
0.17 |
| chr6_124666813_124667013 | 0.12 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
3698 |
0.1 |
| chr14_64865582_64865960 | 0.12 |
Gm37183 |
predicted gene, 37183 |
26512 |
0.14 |
| chr10_8227062_8227243 | 0.12 |
Gm30906 |
predicted gene, 30906 |
51021 |
0.16 |
| chr6_145767466_145768203 | 0.12 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
17263 |
0.18 |
| chr10_69153121_69153293 | 0.12 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
1773 |
0.28 |
| chr14_28450767_28450938 | 0.12 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
4669 |
0.24 |
| chr7_92560754_92560964 | 0.12 |
Ccdc90b |
coiled-coil domain containing 90B |
290 |
0.9 |
| chr12_25817244_25817410 | 0.12 |
Gm9321 |
predicted gene 9321 |
3214 |
0.34 |
| chr4_134606128_134606305 | 0.12 |
Mir6403 |
microRNA 6403 |
38365 |
0.12 |
| chr11_69326130_69326304 | 0.12 |
Kcnab3 |
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
41 |
0.93 |
| chrX_60330664_60330849 | 0.11 |
Mir505 |
microRNA 505 |
63736 |
0.1 |
| chr2_38613882_38614065 | 0.11 |
Gm13586 |
predicted gene 13586 |
8828 |
0.13 |
| chr18_12005324_12005475 | 0.11 |
Tmem241 |
transmembrane protein 241 |
69232 |
0.1 |
| chr9_83599585_83599736 | 0.11 |
Sh3bgrl2 |
SH3 domain binding glutamic acid-rich protein like 2 |
22829 |
0.16 |
| chr7_92769303_92769470 | 0.10 |
Rab30 |
RAB30, member RAS oncogene family |
8197 |
0.14 |
| chr7_112121062_112121238 | 0.10 |
Usp47 |
ubiquitin specific peptidase 47 |
30444 |
0.19 |
| chr7_81368454_81368631 | 0.10 |
Cpeb1 |
cytoplasmic polyadenylation element binding protein 1 |
3899 |
0.19 |
| chr19_23110526_23110678 | 0.10 |
2410080I02Rik |
RIKEN cDNA 2410080I02 gene |
24298 |
0.14 |
| chr15_50015079_50015249 | 0.10 |
Gm49186 |
predicted gene, 49186 |
101782 |
0.09 |
| chr19_7557819_7557970 | 0.10 |
Plaat3 |
phospholipase A and acyltransferase 3 |
409 |
0.82 |
| chr1_23170862_23171024 | 0.10 |
Gm29506 |
predicted gene 29506 |
64730 |
0.09 |
| chr7_19692038_19692206 | 0.10 |
Apoc1 |
apolipoprotein C-I |
369 |
0.68 |
| chr19_34166072_34166246 | 0.10 |
Ankrd22 |
ankyrin repeat domain 22 |
106 |
0.96 |
| chr4_123419872_123420087 | 0.10 |
Macf1 |
microtubule-actin crosslinking factor 1 |
7751 |
0.17 |
| chr17_85040792_85040975 | 0.10 |
Slc3a1 |
solute carrier family 3, member 1 |
12507 |
0.18 |
| chr19_32544319_32544470 | 0.10 |
Gm36419 |
predicted gene, 36419 |
1046 |
0.53 |
| chr3_96561912_96562186 | 0.10 |
Txnip |
thioredoxin interacting protein |
1995 |
0.13 |
| chr11_7519967_7520118 | 0.10 |
Gm11986 |
predicted gene 11986 |
229668 |
0.02 |
| chr3_41580995_41581174 | 0.10 |
Jade1 |
jade family PHD finger 1 |
228 |
0.92 |
| chr2_84752566_84752717 | 0.10 |
Gm19426 |
predicted gene, 19426 |
8986 |
0.08 |
| chr17_30040374_30040525 | 0.10 |
Zfand3 |
zinc finger, AN1-type domain 3 |
19933 |
0.14 |
| chr2_165020771_165020952 | 0.09 |
Ncoa5 |
nuclear receptor coactivator 5 |
2352 |
0.22 |
| chr6_49252322_49252697 | 0.09 |
Gm43980 |
predicted gene, 43980 |
2342 |
0.22 |
| chr18_74810329_74810480 | 0.09 |
Acaa2 |
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
17021 |
0.15 |
| chr5_138080510_138080792 | 0.09 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr2_72866286_72866437 | 0.09 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
52644 |
0.12 |
| chr1_157443382_157443559 | 0.09 |
Cryzl2 |
crystallin zeta like 2 |
15107 |
0.13 |
| chr7_100208092_100208243 | 0.09 |
Gm7067 |
predicted gene 7067 |
9819 |
0.12 |
| chr7_5091742_5091950 | 0.09 |
Epn1 |
epsin 1 |
2008 |
0.13 |
| chr5_87081067_87081299 | 0.09 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
9974 |
0.11 |
| chr4_137377515_137377675 | 0.09 |
2810405F17Rik |
RIKEN cDNA 2810405F17 gene |
10787 |
0.12 |
| chr19_26600149_26600340 | 0.09 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
4806 |
0.21 |
| chr8_93959067_93959388 | 0.09 |
Gnao1 |
guanine nucleotide binding protein, alpha O |
3617 |
0.19 |
| chr2_105263595_105263746 | 0.09 |
Them7 |
thioesterase superfamily member 7 |
39328 |
0.17 |
| chr11_86917162_86917343 | 0.09 |
Ypel2 |
yippee like 2 |
54772 |
0.11 |
| chr2_48441967_48442157 | 0.09 |
Gm13481 |
predicted gene 13481 |
15183 |
0.23 |
| chr2_156204536_156204687 | 0.09 |
Phf20 |
PHD finger protein 20 |
7855 |
0.13 |
| chr1_127011786_127011937 | 0.09 |
Gm5261 |
predicted gene 5261 |
38835 |
0.2 |
| chr4_118143703_118144018 | 0.09 |
Kdm4a |
lysine (K)-specific demethylase 4A |
2899 |
0.21 |
| chr16_23103038_23103338 | 0.09 |
Eif4a2 |
eukaryotic translation initiation factor 4A2 |
4256 |
0.07 |
| chr7_83611767_83612180 | 0.09 |
Tmc3 |
transmembrane channel-like gene family 3 |
14446 |
0.12 |
| chr4_140658235_140658404 | 0.09 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
7693 |
0.15 |
| chr7_6838397_6838557 | 0.09 |
Gm44584 |
predicted gene 44584 |
29476 |
0.1 |
| chr3_132917768_132917922 | 0.09 |
Npnt |
nephronectin |
11486 |
0.16 |
| chr18_10448764_10448942 | 0.09 |
Greb1l |
growth regulation by estrogen in breast cancer-like |
9372 |
0.2 |
| chr13_95664015_95664203 | 0.09 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
1591 |
0.31 |
| chr9_78476442_78477037 | 0.09 |
Gm26377 |
predicted gene, 26377 |
1896 |
0.18 |
| chr14_21563664_21563834 | 0.09 |
Kat6b |
K(lysine) acetyltransferase 6B |
46874 |
0.15 |
| chr8_35433889_35434059 | 0.09 |
Gm34853 |
predicted gene, 34853 |
5760 |
0.18 |
| chr16_14704222_14704423 | 0.09 |
Snai2 |
snail family zinc finger 2 |
1530 |
0.48 |
| chr11_100836746_100836921 | 0.09 |
Stat5b |
signal transducer and activator of transcription 5B |
13705 |
0.12 |
| chr17_63865694_63865894 | 0.08 |
Fer |
fer (fms/fps related) protein kinase |
1779 |
0.35 |
| chr1_21315523_21315674 | 0.08 |
Gm21909 |
predicted gene, 21909 |
722 |
0.5 |
| chr2_49521459_49521660 | 0.08 |
Epc2 |
enhancer of polycomb homolog 2 |
7316 |
0.28 |
| chr6_30212058_30212450 | 0.08 |
Ube2h |
ubiquitin-conjugating enzyme E2H |
31549 |
0.11 |
| chr1_172643169_172643320 | 0.08 |
Dusp23 |
dual specificity phosphatase 23 |
10282 |
0.14 |
| chr2_135297004_135297155 | 0.08 |
Plcb1 |
phospholipase C, beta 1 |
47906 |
0.18 |
| chr1_90968289_90968440 | 0.08 |
Rab17 |
RAB17, member RAS oncogene family |
697 |
0.6 |
| chr10_76590264_76590545 | 0.08 |
Ftcd |
formiminotransferase cyclodeaminase |
3271 |
0.16 |
| chr7_116427721_116427872 | 0.08 |
Pik3c2a |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
15643 |
0.12 |
| chr7_98912975_98913128 | 0.08 |
Gm45187 |
predicted gene 45187 |
7964 |
0.14 |
| chr4_60418791_60418999 | 0.08 |
Mup9 |
major urinary protein 9 |
1689 |
0.29 |
| chr10_99263084_99263251 | 0.08 |
Dusp6 |
dual specificity phosphatase 6 |
64 |
0.95 |
| chr19_38356216_38356572 | 0.08 |
Gm50150 |
predicted gene, 50150 |
13780 |
0.14 |
| chr12_7994294_7994990 | 0.08 |
Apob |
apolipoprotein B |
16863 |
0.21 |
| chr19_6969960_6970168 | 0.08 |
Plcb3 |
phospholipase C, beta 3 |
250 |
0.78 |
| chr17_56643312_56643469 | 0.08 |
Catsperd |
cation channel sperm associated auxiliary subunit delta |
8392 |
0.1 |
| chr13_104035617_104035778 | 0.08 |
Nln |
neurolysin (metallopeptidase M3 family) |
870 |
0.63 |
| chr12_16877456_16877979 | 0.08 |
Rock2 |
Rho-associated coiled-coil containing protein kinase 2 |
17178 |
0.14 |
| chr17_5203438_5203609 | 0.08 |
Gm15599 |
predicted gene 15599 |
91413 |
0.08 |
| chr11_63791140_63791492 | 0.08 |
Gm12288 |
predicted gene 12288 |
26984 |
0.21 |
| chr3_131489084_131489522 | 0.08 |
Sgms2 |
sphingomyelin synthase 2 |
1176 |
0.58 |
| chrX_169892253_169892417 | 0.08 |
Mid1 |
midline 1 |
12671 |
0.24 |
| chr5_130264636_130264798 | 0.08 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
5787 |
0.13 |
| chr10_96230406_96230590 | 0.08 |
4930459C07Rik |
RIKEN cDNA 4930459C07 gene |
4291 |
0.23 |
| chr17_88063910_88064234 | 0.08 |
Fbxo11 |
F-box protein 11 |
815 |
0.66 |
| chr17_56470014_56470183 | 0.08 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
4529 |
0.17 |
| chr8_13676545_13676707 | 0.08 |
Rasa3 |
RAS p21 protein activator 3 |
825 |
0.6 |
| chr12_70777982_70778133 | 0.08 |
Gm40437 |
predicted gene, 40437 |
47288 |
0.12 |
| chr4_6959204_6959408 | 0.08 |
Tox |
thymocyte selection-associated high mobility group box |
31177 |
0.23 |
| chr13_39657075_39657270 | 0.08 |
Gm47352 |
predicted gene, 47352 |
7661 |
0.21 |
| chr4_60218523_60218926 | 0.08 |
Mup8 |
major urinary protein 8 |
3856 |
0.22 |
| chr2_157830426_157830940 | 0.08 |
Ctnnbl1 |
catenin, beta like 1 |
40819 |
0.14 |
| chr19_40415247_40415418 | 0.08 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
13065 |
0.23 |
| chr11_23858437_23858588 | 0.07 |
Papolg |
poly(A) polymerase gamma |
9958 |
0.19 |
| chr3_85813449_85813618 | 0.07 |
Fam160a1 |
family with sequence similarity 160, member A1 |
3758 |
0.23 |
| chr19_58311318_58311494 | 0.07 |
Gm16277 |
predicted gene 16277 |
107267 |
0.07 |
| chr9_116091822_116091994 | 0.07 |
Gm9385 |
predicted pseudogene 9385 |
50844 |
0.13 |
| chr13_104636027_104636417 | 0.07 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
48709 |
0.17 |
| chr6_115476517_115476745 | 0.07 |
Gm44079 |
predicted gene, 44079 |
11641 |
0.17 |
| chr2_165007913_165008284 | 0.07 |
Ncoa5 |
nuclear receptor coactivator 5 |
3435 |
0.18 |
| chr9_100370018_100370211 | 0.07 |
Gm34397 |
predicted gene, 34397 |
14932 |
0.18 |
| chr10_36074127_36074466 | 0.07 |
Gm47048 |
predicted gene, 47048 |
21580 |
0.28 |
| chr12_8395088_8395287 | 0.07 |
Gm48075 |
predicted gene, 48075 |
978 |
0.55 |
| chr3_5213826_5214044 | 0.07 |
Gm10748 |
predicted gene 10748 |
828 |
0.62 |
| chr8_11149308_11149679 | 0.07 |
Gm44717 |
predicted gene 44717 |
247 |
0.91 |
| chr5_119581034_119581457 | 0.07 |
Tbx3os1 |
T-box 3, opposite strand 1 |
6299 |
0.18 |
| chr11_4446565_4446734 | 0.07 |
Hormad2 |
HORMA domain containing 2 |
5544 |
0.21 |
| chr17_57233093_57233442 | 0.07 |
C3 |
complement component 3 |
5131 |
0.12 |
| chr14_47220584_47220749 | 0.07 |
Gm49137 |
predicted gene, 49137 |
5928 |
0.1 |
| chr5_148535815_148535974 | 0.07 |
Ubl3 |
ubiquitin-like 3 |
1696 |
0.37 |
| chr11_32681086_32681471 | 0.07 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
38374 |
0.16 |
| chr16_24228826_24228998 | 0.07 |
Gm31814 |
predicted gene, 31814 |
12388 |
0.21 |
| chr10_87546435_87546600 | 0.07 |
Pah |
phenylalanine hydroxylase |
156 |
0.96 |
| chr15_61055209_61055425 | 0.07 |
Gm38563 |
predicted gene, 38563 |
102552 |
0.06 |
| chr5_87003545_87003733 | 0.07 |
Ugt2b35 |
UDP glucuronosyltransferase 2 family, polypeptide B35 |
2779 |
0.14 |
| chr6_112531173_112531324 | 0.07 |
Gm27944 |
predicted gene, 27944 |
17801 |
0.15 |
| chr2_71499002_71499251 | 0.07 |
Gm23253 |
predicted gene, 23253 |
3855 |
0.16 |
| chr18_47503988_47504152 | 0.07 |
Gm50223 |
predicted gene, 50223 |
26566 |
0.16 |
| chr19_23070301_23070453 | 0.07 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
5476 |
0.2 |
| chr2_52498942_52499094 | 0.07 |
Gm13541 |
predicted gene 13541 |
52951 |
0.11 |
| chr4_60659172_60659323 | 0.07 |
Mup11 |
major urinary protein 11 |
491 |
0.78 |
| chr8_14259594_14259775 | 0.07 |
Gm26184 |
predicted gene, 26184 |
13627 |
0.23 |
| chr1_84877752_84878205 | 0.07 |
Fbxo36 |
F-box protein 36 |
8827 |
0.16 |
| chr15_3601373_3601655 | 0.07 |
Gm22031 |
predicted gene, 22031 |
1883 |
0.41 |
| chr19_36735506_36735662 | 0.07 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
1069 |
0.56 |
| chr8_79518919_79519070 | 0.07 |
Gm8077 |
predicted gene 8077 |
20816 |
0.14 |
| chr14_8326780_8326935 | 0.07 |
Fam107a |
family with sequence similarity 107, member A |
8834 |
0.19 |
| chr3_146617776_146618365 | 0.07 |
Dnase2b |
deoxyribonuclease II beta |
2474 |
0.2 |
| chr6_116167704_116167874 | 0.07 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
3908 |
0.16 |
| chr19_33078714_33079266 | 0.07 |
Gm29863 |
predicted gene, 29863 |
8918 |
0.23 |
| chr9_21960198_21960357 | 0.07 |
Epor |
erythropoietin receptor |
2287 |
0.14 |
| chr7_3145683_3146014 | 0.07 |
Gm29708 |
predicted gene, 29708 |
4143 |
0.09 |
| chr19_4616707_4616858 | 0.07 |
Lrfn4 |
leucine rich repeat and fibronectin type III domain containing 4 |
1115 |
0.34 |
| chr19_61057720_61057890 | 0.07 |
Gm22520 |
predicted gene, 22520 |
44260 |
0.12 |
| chr2_169724086_169724281 | 0.07 |
Tshz2 |
teashirt zinc finger family member 2 |
90507 |
0.08 |
| chr3_27677723_27677906 | 0.07 |
Fndc3b |
fibronectin type III domain containing 3B |
32584 |
0.21 |
| chr5_33070400_33070569 | 0.07 |
Rpl35a-ps5 |
ribosomal protein L35A, pseudogene 5 |
5554 |
0.15 |
| chr12_40596811_40597378 | 0.07 |
Dock4 |
dedicator of cytokinesis 4 |
150758 |
0.04 |
| chr4_45967836_45967998 | 0.07 |
Tdrd7 |
tudor domain containing 7 |
2583 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |