Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
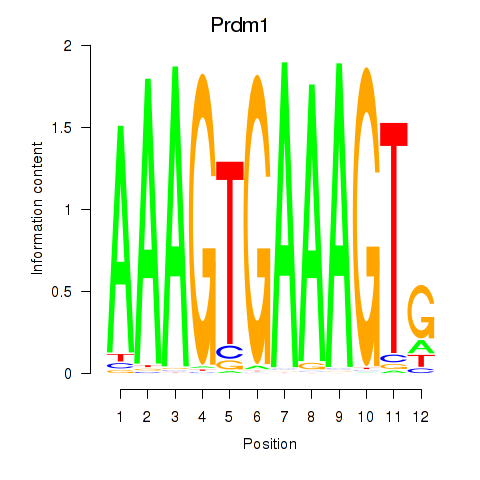
Results for Prdm1
Z-value: 1.86
Transcription factors associated with Prdm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm1
|
ENSMUSG00000038151.6 | PR domain containing 1, with ZNF domain |
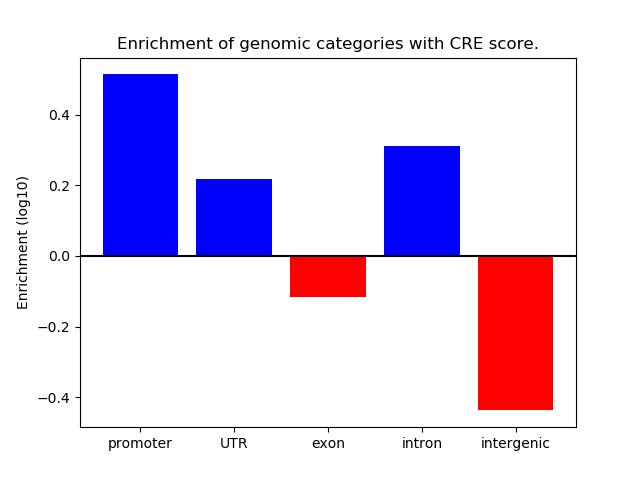
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_44454950_44455114 | Prdm1 | 2178 | 0.316618 | 0.71 | 1.2e-01 | Click! |
| chr10_44474418_44474575 | Prdm1 | 15748 | 0.184920 | 0.58 | 2.2e-01 | Click! |
| chr10_44481546_44481706 | Prdm1 | 22878 | 0.169199 | 0.47 | 3.4e-01 | Click! |
| chr10_44455197_44455348 | Prdm1 | 1938 | 0.343142 | -0.39 | 4.5e-01 | Click! |
| chr10_44447565_44447742 | Prdm1 | 9557 | 0.192420 | 0.35 | 4.9e-01 | Click! |
Activity of the Prdm1 motif across conditions
Conditions sorted by the z-value of the Prdm1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
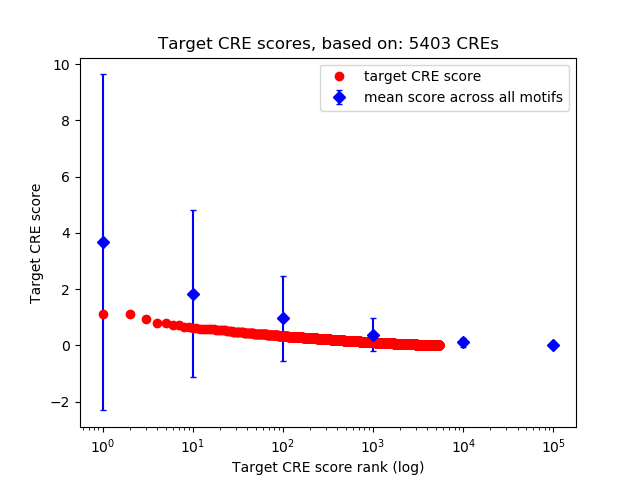
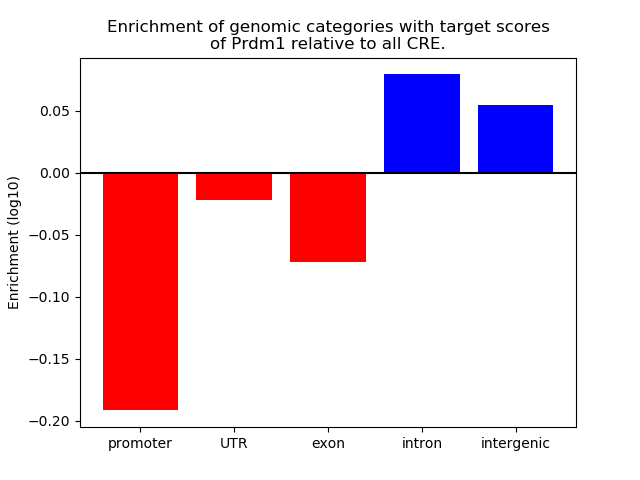
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_145775927_145776137 | 1.12 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
15284 |
0.2 |
| chr18_7657901_7658087 | 1.10 |
Mpp7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
31128 |
0.17 |
| chr19_26823384_26823535 | 0.93 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
448 |
0.84 |
| chr11_5905326_5905477 | 0.79 |
Gck |
glucokinase |
3314 |
0.14 |
| chr3_96883833_96883984 | 0.78 |
Gpr89 |
G protein-coupled receptor 89 |
7700 |
0.13 |
| chr7_84457901_84458052 | 0.74 |
Gm45175 |
predicted gene 45175 |
13425 |
0.16 |
| chr7_72323573_72323742 | 0.71 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
17049 |
0.28 |
| chr6_117577757_117577908 | 0.65 |
Gm9946 |
predicted gene 9946 |
10160 |
0.2 |
| chr1_178600841_178601326 | 0.64 |
Gm24405 |
predicted gene, 24405 |
35822 |
0.18 |
| chr13_41386761_41386912 | 0.62 |
Gm48571 |
predicted gene, 48571 |
6880 |
0.15 |
| chr6_43850496_43850673 | 0.60 |
Gm7783 |
predicted gene 7783 |
6320 |
0.26 |
| chr15_3468057_3468222 | 0.59 |
Ghr |
growth hormone receptor |
3505 |
0.35 |
| chr11_16528502_16528653 | 0.58 |
Akt2-ps |
thymoma viral proto-oncogene 2, pseudogene |
14044 |
0.2 |
| chr9_74892937_74893252 | 0.58 |
Onecut1 |
one cut domain, family member 1 |
26610 |
0.14 |
| chr6_121171179_121171645 | 0.58 |
Pex26 |
peroxisomal biogenesis factor 26 |
12255 |
0.13 |
| chr3_111344475_111344661 | 0.57 |
Gm9314 |
predicted gene 9314 |
21711 |
0.29 |
| chr2_21117164_21117329 | 0.57 |
Gm13376 |
predicted gene 13376 |
20147 |
0.17 |
| chr13_112902308_112902475 | 0.55 |
Mtrex |
Mtr4 exosome RNA helicase |
15283 |
0.12 |
| chr3_69139928_69140109 | 0.55 |
Gm1647 |
predicted gene 1647 |
8785 |
0.14 |
| chr12_104342354_104343298 | 0.54 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
4340 |
0.13 |
| chr9_114621205_114621361 | 0.54 |
Cnot10 |
CCR4-NOT transcription complex, subunit 10 |
18680 |
0.14 |
| chr11_88926781_88926936 | 0.54 |
Scpep1os |
serine carboxypeptidase 1, opposite strand |
20930 |
0.1 |
| chr15_37998302_37998493 | 0.53 |
Ubr5 |
ubiquitin protein ligase E3 component n-recognin 5 |
1730 |
0.31 |
| chr16_4211280_4211431 | 0.52 |
Crebbp |
CREB binding protein |
2058 |
0.29 |
| chr2_134502496_134502663 | 0.52 |
Hao1 |
hydroxyacid oxidase 1, liver |
51728 |
0.18 |
| chr6_145863813_145864067 | 0.51 |
Gm43909 |
predicted gene, 43909 |
643 |
0.6 |
| chr4_148616552_148616759 | 0.50 |
Tardbp |
TAR DNA binding protein |
325 |
0.81 |
| chr9_71694307_71694467 | 0.49 |
Cgnl1 |
cingulin-like 1 |
46037 |
0.14 |
| chr8_128078877_128079030 | 0.48 |
Mir21c |
microRNA 21c |
199272 |
0.03 |
| chr11_26413799_26413986 | 0.48 |
Gm12713 |
predicted gene 12713 |
4685 |
0.26 |
| chr12_30344893_30345053 | 0.48 |
Sntg2 |
syntrophin, gamma 2 |
12710 |
0.25 |
| chr19_36932520_36932671 | 0.47 |
Btaf1 |
B-TFIID TATA-box binding protein associated factor 1 |
6516 |
0.21 |
| chr10_68137101_68137496 | 0.47 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
672 |
0.78 |
| chr1_182100464_182100642 | 0.47 |
Srp9 |
signal recognition particle 9 |
24184 |
0.13 |
| chr16_34921418_34921569 | 0.46 |
Mylk |
myosin, light polypeptide kinase |
8849 |
0.19 |
| chr3_107238884_107239100 | 0.46 |
Prok1 |
prokineticin 1 |
340 |
0.83 |
| chr1_69634374_69634531 | 0.45 |
Gm28112 |
predicted gene 28112 |
27609 |
0.17 |
| chr17_63181800_63181984 | 0.45 |
Gm24730 |
predicted gene, 24730 |
20846 |
0.24 |
| chr11_80103009_80103182 | 0.45 |
Atad5 |
ATPase family, AAA domain containing 5 |
92 |
0.96 |
| chr16_43169614_43169782 | 0.44 |
Gm15712 |
predicted gene 15712 |
14875 |
0.21 |
| chr11_16900094_16900245 | 0.44 |
Egfr |
epidermal growth factor receptor |
5016 |
0.22 |
| chr12_80932956_80933128 | 0.44 |
1700052I22Rik |
RIKEN cDNA 1700052I22 gene |
8593 |
0.13 |
| chr6_72000258_72000422 | 0.44 |
Gm26628 |
predicted gene, 26628 |
36565 |
0.1 |
| chr9_74304589_74304740 | 0.43 |
Wdr72 |
WD repeat domain 72 |
30530 |
0.19 |
| chr19_38419073_38419264 | 0.43 |
Slc35g1 |
solute carrier family 35, member G1 |
23127 |
0.14 |
| chr3_52458834_52458985 | 0.43 |
Gm38098 |
predicted gene, 38098 |
33501 |
0.17 |
| chr12_69768209_69768388 | 0.42 |
Mir681 |
microRNA 681 |
4354 |
0.15 |
| chr2_134853625_134853800 | 0.42 |
Gm14036 |
predicted gene 14036 |
49763 |
0.15 |
| chr12_104082595_104082890 | 0.42 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2093 |
0.18 |
| chr7_79275538_79275706 | 0.41 |
Gm44639 |
predicted gene 44639 |
2145 |
0.2 |
| chr16_49775651_49775805 | 0.41 |
Gm15518 |
predicted gene 15518 |
23142 |
0.2 |
| chr12_110044398_110044582 | 0.41 |
Gm47187 |
predicted gene, 47187 |
17780 |
0.11 |
| chr4_6262292_6262457 | 0.41 |
Gm11798 |
predicted gene 11798 |
1409 |
0.43 |
| chr3_79994302_79994453 | 0.41 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
60515 |
0.11 |
| chr5_151018980_151019139 | 0.41 |
Gm8675 |
predicted gene 8675 |
9758 |
0.23 |
| chr4_3751092_3751243 | 0.40 |
Lyn |
LYN proto-oncogene, Src family tyrosine kinase |
5290 |
0.17 |
| chr2_70666883_70667034 | 0.40 |
Gorasp2 |
golgi reassembly stacking protein 2 |
4758 |
0.18 |
| chr2_122255801_122256080 | 0.40 |
Sord |
sorbitol dehydrogenase |
21191 |
0.09 |
| chr17_6867831_6868002 | 0.40 |
Mir692-1 |
microRNA 692-1 |
27421 |
0.12 |
| chr17_57198725_57198876 | 0.40 |
Tnfsf14 |
tumor necrosis factor (ligand) superfamily, member 14 |
4623 |
0.13 |
| chr1_39368822_39369001 | 0.40 |
Rpl31 |
ribosomal protein L31 |
762 |
0.58 |
| chr1_21829923_21830095 | 0.39 |
Gm38243 |
predicted gene, 38243 |
10505 |
0.24 |
| chr2_52613475_52613626 | 0.39 |
Bloc1s2-ps |
biogenesis of lysosomal organelles complex-1, subunit 2, pseudogene |
6204 |
0.24 |
| chr6_95018081_95018258 | 0.39 |
4930511E03Rik |
RIKEN cDNA 4930511E03 gene |
74335 |
0.09 |
| chr15_59141725_59141916 | 0.39 |
Mtss1 |
MTSS I-BAR domain containing 1 |
59815 |
0.11 |
| chr4_155071078_155071229 | 0.39 |
Pex10 |
peroxisomal biogenesis factor 10 |
4094 |
0.15 |
| chr2_24040580_24040761 | 0.38 |
Hnmt |
histamine N-methyltransferase |
8196 |
0.23 |
| chr6_23080148_23080299 | 0.38 |
Ptprz1 |
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
31951 |
0.15 |
| chr12_103991999_103992602 | 0.38 |
Serpina11 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
2343 |
0.16 |
| chr10_121571625_121571789 | 0.38 |
Tbk1 |
TANK-binding kinase 1 |
3226 |
0.18 |
| chr11_55468108_55468290 | 0.38 |
G3bp1 |
GTPase activating protein (SH3 domain) binding protein 1 |
1486 |
0.25 |
| chr13_74505877_74506069 | 0.38 |
Gm49763 |
predicted gene, 49763 |
11830 |
0.1 |
| chr5_90315666_90315817 | 0.37 |
Ankrd17 |
ankyrin repeat domain 17 |
11637 |
0.21 |
| chr5_51539672_51539839 | 0.37 |
Ppargc1a |
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
14116 |
0.17 |
| chr7_71232911_71233088 | 0.37 |
6030442E23Rik |
RIKEN cDNA 6030442E23 gene |
15950 |
0.2 |
| chr14_7832222_7832456 | 0.37 |
Flnb |
filamin, beta |
14382 |
0.16 |
| chr18_14825706_14825857 | 0.36 |
Gm21055 |
predicted gene, 21055 |
4887 |
0.25 |
| chr1_172590517_172590678 | 0.36 |
Slamf8 |
SLAM family member 8 |
29 |
0.96 |
| chr19_55793163_55793324 | 0.36 |
Ppnr |
per-pentamer repeat gene |
48429 |
0.16 |
| chr4_3373115_3373279 | 0.36 |
Gm11784 |
predicted gene 11784 |
13649 |
0.2 |
| chr9_122848440_122848788 | 0.36 |
Gm47140 |
predicted gene, 47140 |
196 |
0.89 |
| chr7_4655984_4656315 | 0.36 |
Gm44878 |
predicted gene 44878 |
2511 |
0.11 |
| chr15_53724232_53724383 | 0.35 |
1700015H07Rik |
RIKEN cDNA 1700015H07 gene |
102652 |
0.07 |
| chr17_86150939_86151254 | 0.35 |
Srbd1 |
S1 RNA binding domain 1 |
5921 |
0.21 |
| chr16_56038236_56038391 | 0.35 |
Trmt10c |
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
494 |
0.65 |
| chr3_83663805_83664183 | 0.35 |
Rpl28-ps2 |
ribosomal protein L28, pseudogene 2 |
22727 |
0.19 |
| chr8_76540217_76540646 | 0.35 |
Gm27355 |
predicted gene, 27355 |
78851 |
0.1 |
| chr3_98743356_98743926 | 0.35 |
Hsd3b3 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
9824 |
0.13 |
| chr4_49542856_49543140 | 0.35 |
Aldob |
aldolase B, fructose-bisphosphate |
3112 |
0.18 |
| chr14_116240919_116241084 | 0.34 |
Gm20713 |
predicted gene 20713 |
369723 |
0.01 |
| chr4_3724826_3724987 | 0.34 |
Gm11805 |
predicted gene 11805 |
3880 |
0.2 |
| chr14_95884398_95884549 | 0.34 |
Spertl |
spermatid associated like |
3207 |
0.33 |
| chr6_146903752_146903913 | 0.34 |
Ppfibp1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
15243 |
0.15 |
| chr6_114879701_114879922 | 0.34 |
Vgll4 |
vestigial like family member 4 |
4704 |
0.23 |
| chr5_100557088_100557255 | 0.34 |
Plac8 |
placenta-specific 8 |
5687 |
0.14 |
| chr6_38506694_38506867 | 0.34 |
Mir7670 |
microRNA 7670 |
545 |
0.71 |
| chr3_30105629_30105802 | 0.34 |
Mecom |
MDS1 and EVI1 complex locus |
34708 |
0.17 |
| chr16_21836755_21836930 | 0.34 |
Gm49607 |
predicted gene, 49607 |
287 |
0.85 |
| chr11_16777068_16777446 | 0.33 |
Egfr |
epidermal growth factor receptor |
25027 |
0.17 |
| chr9_71175821_71176011 | 0.33 |
Gm32511 |
predicted gene, 32511 |
7140 |
0.18 |
| chr13_81330589_81330889 | 0.33 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
12097 |
0.26 |
| chr2_109714999_109715174 | 0.32 |
Bdnf |
brain derived neurotrophic factor |
5703 |
0.22 |
| chr12_118296347_118296498 | 0.32 |
Sp4 |
trans-acting transcription factor 4 |
4946 |
0.29 |
| chr3_18157205_18157356 | 0.32 |
Gm23686 |
predicted gene, 23686 |
20345 |
0.21 |
| chr18_84210867_84211018 | 0.32 |
Gm50311 |
predicted gene, 50311 |
8018 |
0.25 |
| chr14_66027915_66028101 | 0.32 |
Gm10233 |
predicted pseudogene 10233 |
17072 |
0.14 |
| chr8_84650202_84650366 | 0.32 |
Cacna1a |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
11293 |
0.1 |
| chr6_54465003_54465192 | 0.32 |
Wipf3 |
WAS/WASL interacting protein family, member 3 |
6905 |
0.17 |
| chr1_177794963_177795127 | 0.32 |
Adss |
adenylosuccinate synthetase, non muscle |
1456 |
0.38 |
| chr9_55281590_55281872 | 0.32 |
Nrg4 |
neuregulin 4 |
1841 |
0.33 |
| chr6_38868049_38868218 | 0.32 |
Tbxas1 |
thromboxane A synthase 1, platelet |
7271 |
0.21 |
| chr11_46309219_46309377 | 0.31 |
Cyfip2 |
cytoplasmic FMR1 interacting protein 2 |
2922 |
0.22 |
| chr11_54413058_54413209 | 0.31 |
Gm12225 |
predicted gene 12225 |
8993 |
0.13 |
| chr7_130059814_130059979 | 0.31 |
Gm23847 |
predicted gene, 23847 |
24802 |
0.24 |
| chr8_14996937_14997102 | 0.31 |
Arhgef10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
883 |
0.45 |
| chr2_114610179_114610347 | 0.31 |
Gm13972 |
predicted gene 13972 |
44162 |
0.16 |
| chr19_37213872_37214035 | 0.31 |
Marchf5 |
membrane associated ring-CH-type finger 5 |
3237 |
0.17 |
| chr17_42887528_42887679 | 0.31 |
Cd2ap |
CD2-associated protein |
10938 |
0.28 |
| chr6_120917276_120917440 | 0.31 |
Bid |
BH3 interacting domain death agonist |
505 |
0.67 |
| chr17_8343757_8343973 | 0.31 |
Prr18 |
proline rich 18 |
3126 |
0.15 |
| chr9_65809859_65810043 | 0.31 |
Zfp609 |
zinc finger protein 609 |
12039 |
0.14 |
| chr3_95324812_95324966 | 0.31 |
Cers2 |
ceramide synthase 2 |
3949 |
0.1 |
| chr18_43129253_43129421 | 0.31 |
Gm8181 |
predicted gene 8181 |
41822 |
0.12 |
| chr8_11622585_11622761 | 0.30 |
Ankrd10 |
ankyrin repeat domain 10 |
1953 |
0.2 |
| chr8_77241292_77241473 | 0.30 |
Gm45406 |
predicted gene 45406 |
41479 |
0.13 |
| chr5_100418951_100419124 | 0.30 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
2803 |
0.2 |
| chr11_25838781_25838932 | 0.30 |
5730522E02Rik |
RIKEN cDNA 5730522E02 gene |
69715 |
0.13 |
| chr2_118448093_118448276 | 0.30 |
Eif2ak4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
8976 |
0.16 |
| chr15_53168686_53168859 | 0.30 |
Ext1 |
exostosin glycosyltransferase 1 |
27483 |
0.26 |
| chr14_13625050_13625231 | 0.30 |
Sntn |
sentan, cilia apical structure protein |
45736 |
0.17 |
| chr13_62866069_62866220 | 0.30 |
Fbp1 |
fructose bisphosphatase 1 |
6614 |
0.13 |
| chr1_62719851_62720045 | 0.30 |
Gm29083 |
predicted gene 29083 |
1875 |
0.33 |
| chr5_4024477_4024646 | 0.30 |
Akap9 |
A kinase (PRKA) anchor protein (yotiao) 9 |
1146 |
0.5 |
| chr5_98992026_98992185 | 0.30 |
Prkg2 |
protein kinase, cGMP-dependent, type II |
25349 |
0.2 |
| chr10_94545587_94545808 | 0.30 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
266 |
0.92 |
| chr8_86735912_86736063 | 0.30 |
Gm10638 |
predicted gene 10638 |
9466 |
0.17 |
| chr10_90273418_90273600 | 0.30 |
Gm5780 |
predicted gene 5780 |
138692 |
0.05 |
| chr1_157247495_157247658 | 0.29 |
Rasal2 |
RAS protein activator like 2 |
3086 |
0.3 |
| chr2_163670938_163671176 | 0.29 |
Pkig |
protein kinase inhibitor, gamma |
9558 |
0.13 |
| chr6_134544187_134544824 | 0.29 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
2791 |
0.27 |
| chr8_46499145_46499431 | 0.29 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
6456 |
0.17 |
| chr18_65837283_65837437 | 0.29 |
Gm50174 |
predicted gene, 50174 |
73 |
0.94 |
| chr11_16913067_16913239 | 0.29 |
Egfr |
epidermal growth factor receptor |
7968 |
0.19 |
| chr6_51458240_51458453 | 0.29 |
Hnrnpa2b1 |
heterogeneous nuclear ribonucleoprotein A2/B1 |
4838 |
0.18 |
| chr12_58214814_58214997 | 0.29 |
Sstr1 |
somatostatin receptor 1 |
3101 |
0.36 |
| chr17_28437280_28437431 | 0.29 |
Fkbp5 |
FK506 binding protein 5 |
3655 |
0.13 |
| chr2_72114198_72114385 | 0.29 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
26420 |
0.18 |
| chr14_17695082_17695233 | 0.29 |
Thrb |
thyroid hormone receptor beta |
34261 |
0.22 |
| chr11_16769725_16769905 | 0.29 |
Egfr |
epidermal growth factor receptor |
17585 |
0.19 |
| chrX_48054599_48054771 | 0.29 |
Apln |
apelin |
19832 |
0.2 |
| chr15_39111986_39112168 | 0.29 |
Slc25a32 |
solute carrier family 25, member 32 |
555 |
0.51 |
| chr16_95579480_95579639 | 0.29 |
Erg |
ETS transcription factor |
7034 |
0.29 |
| chr11_118211857_118212019 | 0.29 |
Cyth1 |
cytohesin 1 |
804 |
0.57 |
| chr2_38108700_38108871 | 0.28 |
Gm44291 |
predicted gene, 44291 |
1489 |
0.45 |
| chr19_58421265_58421439 | 0.28 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
33114 |
0.19 |
| chr10_121501382_121501572 | 0.28 |
Gm46204 |
predicted gene, 46204 |
3874 |
0.13 |
| chr4_70521535_70521760 | 0.28 |
Megf9 |
multiple EGF-like-domains 9 |
13281 |
0.3 |
| chr15_85675723_85675883 | 0.28 |
Lncppara |
long noncoding RNA near Ppara |
22187 |
0.12 |
| chr10_79769428_79769606 | 0.28 |
Rnf126 |
ring finger protein 126 |
2565 |
0.11 |
| chr10_120015234_120015396 | 0.28 |
Grip1 |
glutamate receptor interacting protein 1 |
22209 |
0.22 |
| chr2_121439848_121440228 | 0.28 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
1113 |
0.25 |
| chr9_105746936_105747087 | 0.28 |
Gm38314 |
predicted gene, 38314 |
19226 |
0.18 |
| chr18_36571831_36571982 | 0.28 |
Ankhd1 |
ankyrin repeat and KH domain containing 1 |
9171 |
0.13 |
| chr1_9578333_9578671 | 0.28 |
Gm6161 |
predicted gene 6161 |
3954 |
0.16 |
| chr8_125346149_125346401 | 0.28 |
Gm16237 |
predicted gene 16237 |
118952 |
0.06 |
| chr3_66122973_66123139 | 0.28 |
Tpi-rs2 |
triosephosphate isomerase related sequence 2 |
6667 |
0.14 |
| chr4_84540692_84540849 | 0.28 |
Bnc2 |
basonuclin 2 |
5520 |
0.32 |
| chr10_87920381_87920532 | 0.28 |
Tyms-ps |
thymidylate synthase, pseudogene |
46391 |
0.12 |
| chr10_123118441_123118756 | 0.28 |
Usp15 |
ubiquitin specific peptidase 15 |
883 |
0.61 |
| chr19_22642267_22642418 | 0.28 |
Trpm3 |
transient receptor potential cation channel, subfamily M, member 3 |
50259 |
0.17 |
| chr14_11691642_11691807 | 0.28 |
Gm48602 |
predicted gene, 48602 |
76219 |
0.1 |
| chr4_150427704_150427878 | 0.27 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
21397 |
0.21 |
| chr6_141655199_141655350 | 0.27 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
3019 |
0.35 |
| chr1_191180455_191180648 | 0.27 |
Atf3 |
activating transcription factor 3 |
2557 |
0.2 |
| chr16_52416810_52416986 | 0.27 |
Alcam |
activated leukocyte cell adhesion molecule |
35567 |
0.23 |
| chr4_108055323_108055845 | 0.27 |
Scp2 |
sterol carrier protein 2, liver |
7705 |
0.14 |
| chr8_78031019_78031191 | 0.27 |
Gm29895 |
predicted gene, 29895 |
12905 |
0.26 |
| chr14_12072661_12072824 | 0.27 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
18418 |
0.23 |
| chr4_46863636_46863790 | 0.27 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
3811 |
0.32 |
| chr15_53297534_53297744 | 0.27 |
Ext1 |
exostosin glycosyltransferase 1 |
48020 |
0.18 |
| chr10_127632738_127633305 | 0.27 |
Gm48815 |
predicted gene, 48815 |
3366 |
0.11 |
| chr11_16902546_16902715 | 0.27 |
Egfr |
epidermal growth factor receptor |
2555 |
0.28 |
| chr8_114152687_114152971 | 0.27 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
19187 |
0.25 |
| chr13_38090008_38090167 | 0.27 |
Gm27387 |
predicted gene, 27387 |
3885 |
0.2 |
| chr10_38970068_38970231 | 0.27 |
Lama4 |
laminin, alpha 4 |
4477 |
0.27 |
| chr9_21969411_21969573 | 0.27 |
Rgl3 |
ral guanine nucleotide dissociation stimulator-like 3 |
4791 |
0.09 |
| chr12_69513282_69513448 | 0.27 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
37815 |
0.11 |
| chr2_48333003_48333182 | 0.27 |
Gm13482 |
predicted gene 13482 |
34184 |
0.18 |
| chr10_39417755_39417908 | 0.27 |
Fyn |
Fyn proto-oncogene |
2178 |
0.32 |
| chr13_4468002_4468176 | 0.27 |
Gm48010 |
predicted gene, 48010 |
1523 |
0.38 |
| chr14_34488757_34488916 | 0.26 |
Gm17110 |
predicted gene 17110 |
12879 |
0.13 |
| chr16_91441296_91441472 | 0.26 |
Gm46562 |
predicted gene, 46562 |
17037 |
0.09 |
| chr13_80890366_80890663 | 0.26 |
Arrdc3 |
arrestin domain containing 3 |
4 |
0.97 |
| chr6_11961160_11961311 | 0.26 |
Phf14 |
PHD finger protein 14 |
25829 |
0.23 |
| chr11_100526464_100526691 | 0.26 |
Acly |
ATP citrate lyase |
1292 |
0.28 |
| chr2_145704854_145705052 | 0.26 |
Gm11763 |
predicted gene 11763 |
3228 |
0.26 |
| chr4_135989135_135989286 | 0.26 |
Pithd1 |
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
1946 |
0.16 |
| chr10_69923910_69924142 | 0.26 |
Ank3 |
ankyrin 3, epithelial |
1469 |
0.57 |
| chr16_31705299_31705479 | 0.26 |
Dlg1 |
discs large MAGUK scaffold protein 1 |
21123 |
0.13 |
| chr10_4105736_4105887 | 0.26 |
Gm25515 |
predicted gene, 25515 |
1498 |
0.45 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.7 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.3 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.2 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.1 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 | 0.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.1 | 0.2 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.1 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.2 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.0 | GO:1903624 | regulation of DNA catabolic process(GO:1903624) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0072711 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.1 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.0 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:2000391 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0034115 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.0 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.0 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.0 | 0.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0043723 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:1904680 | peptide transmembrane transporter activity(GO:1904680) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.0 | GO:0016530 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |