Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
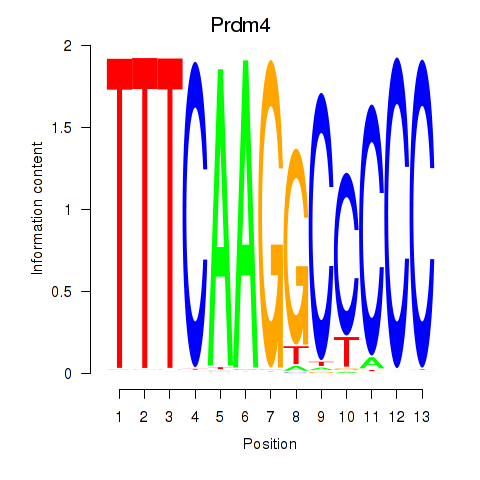
Results for Prdm4
Z-value: 0.63
Transcription factors associated with Prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm4
|
ENSMUSG00000035529.9 | PR domain containing 4 |
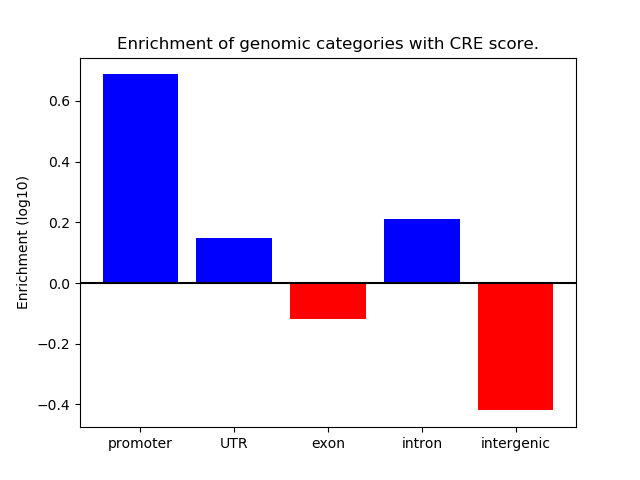
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_85917049_85917211 | Prdm4 | 8 | 0.961476 | -0.71 | 1.1e-01 | Click! |
| chr10_85916562_85916789 | Prdm4 | 19 | 0.961021 | 0.06 | 9.1e-01 | Click! |
Activity of the Prdm4 motif across conditions
Conditions sorted by the z-value of the Prdm4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
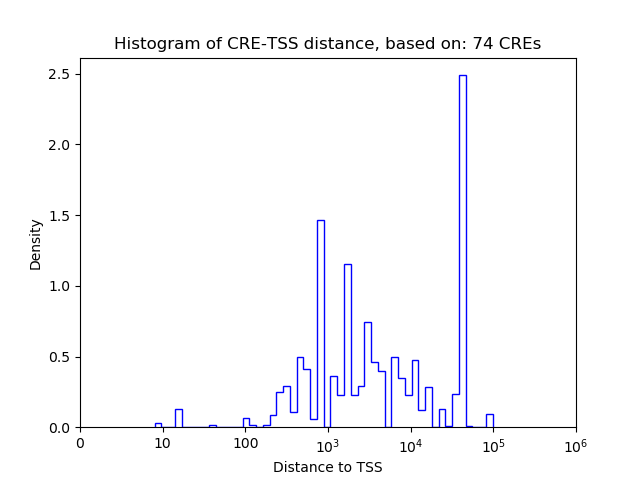
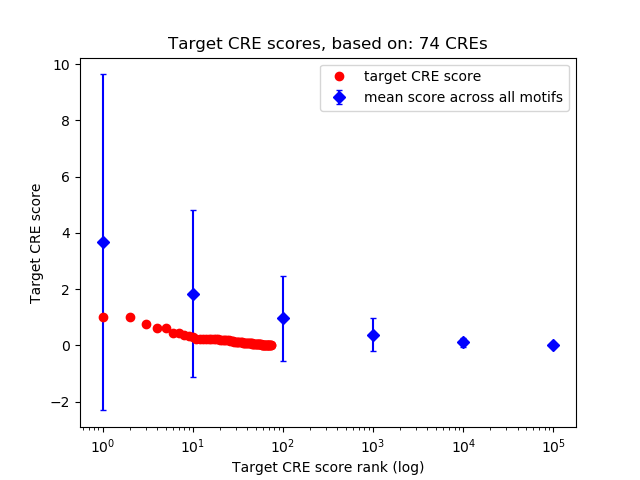
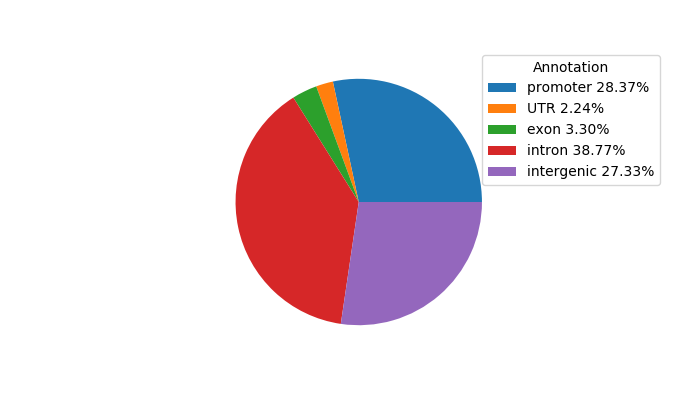
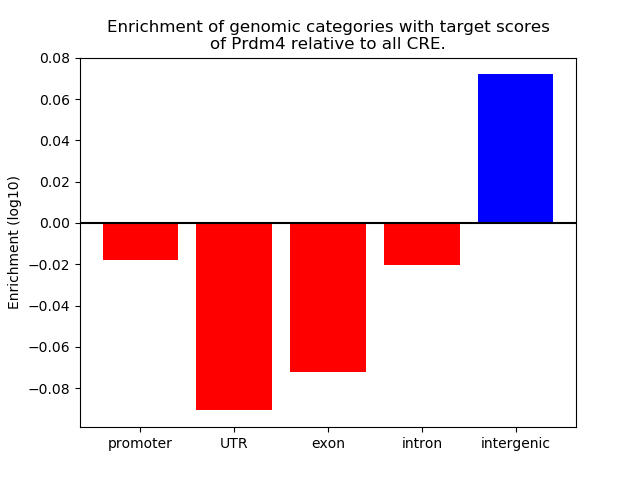
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_40146515_40146666 | 1.00 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
40696 |
0.11 |
| chr17_25866831_25867238 | 1.00 |
Mcrip2 |
MAPK regulated corepressor interacting protein 2 |
802 |
0.3 |
| chr2_34486208_34486368 | 0.75 |
Mapkap1 |
mitogen-activated protein kinase associated protein 1 |
41941 |
0.13 |
| chr3_27752082_27752234 | 0.63 |
Fndc3b |
fibronectin type III domain containing 3B |
40851 |
0.19 |
| chr13_101698805_101699022 | 0.60 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
3191 |
0.3 |
| chr13_69582949_69583107 | 0.45 |
Srd5a1 |
steroid 5 alpha-reductase 1 |
11801 |
0.12 |
| chr17_25476016_25476187 | 0.44 |
Tekt4 |
tektin 4 |
1797 |
0.21 |
| chr19_12635033_12635481 | 0.35 |
Glyat |
glycine-N-acyltransferase |
1724 |
0.21 |
| chr13_69637525_69637701 | 0.32 |
Nsun2 |
NOL1/NOP2/Sun domain family member 2 |
6873 |
0.11 |
| chr13_119617412_119617581 | 0.31 |
Gm48265 |
predicted gene, 48265 |
4024 |
0.15 |
| chr11_32226143_32226339 | 0.24 |
Mpg |
N-methylpurine-DNA glycosylase |
264 |
0.84 |
| chr15_59194800_59194984 | 0.24 |
Rpl7-ps8 |
ribosomal protein L7, pseudogene 8 |
16016 |
0.19 |
| chr3_83025445_83025870 | 0.23 |
Fga |
fibrinogen alpha chain |
419 |
0.79 |
| chrX_150585229_150585420 | 0.23 |
Apex2 |
apurinic/apyrimidinic endonuclease 2 |
883 |
0.5 |
| chr1_172310436_172310610 | 0.22 |
Igsf8 |
immunoglobulin superfamily, member 8 |
1248 |
0.29 |
| chr11_74672141_74672525 | 0.22 |
Cluh |
clustered mitochondria (cluA/CLU1) homolog |
7851 |
0.16 |
| chr12_80111390_80111729 | 0.22 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
1435 |
0.29 |
| chr11_98922376_98922532 | 0.21 |
Cdc6 |
cell division cycle 6 |
1988 |
0.19 |
| chr5_127611544_127611892 | 0.21 |
Slc15a4 |
solute carrier family 15, member 4 |
2367 |
0.25 |
| chr12_51377705_51378103 | 0.19 |
Scfd1 |
Sec1 family domain containing 1 |
312 |
0.91 |
| chr9_108797332_108797857 | 0.19 |
Ip6k2 |
inositol hexaphosphate kinase 2 |
593 |
0.53 |
| chr15_103364813_103364984 | 0.18 |
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
1850 |
0.21 |
| chr7_30920552_30920938 | 0.17 |
Hamp2 |
hepcidin antimicrobial peptide 2 |
3436 |
0.08 |
| chr8_84201590_84201802 | 0.17 |
Gm37352 |
predicted gene, 37352 |
827 |
0.3 |
| chr1_66701224_66701579 | 0.17 |
Rpe |
ribulose-5-phosphate-3-epimerase |
494 |
0.72 |
| chr7_30940285_30940698 | 0.16 |
Hamp |
hepcidin antimicrobial peptide |
3541 |
0.08 |
| chr4_98108774_98108954 | 0.15 |
Gm12691 |
predicted gene 12691 |
37735 |
0.2 |
| chr11_77348689_77348937 | 0.14 |
Ssh2 |
slingshot protein phosphatase 2 |
525 |
0.79 |
| chr13_63241835_63242032 | 0.13 |
Aopep |
aminopeptidase O |
1825 |
0.19 |
| chr10_85102143_85102819 | 0.12 |
Fhl4 |
four and a half LIM domains 4 |
14 |
0.65 |
| chrX_135571644_135571812 | 0.12 |
Nxf7 |
nuclear RNA export factor 7 |
14405 |
0.12 |
| chr2_163556084_163556253 | 0.11 |
Hnf4a |
hepatic nuclear factor 4, alpha |
6085 |
0.14 |
| chr15_36580514_36580708 | 0.11 |
Gm44310 |
predicted gene, 44310 |
2933 |
0.19 |
| chr17_68176889_68177063 | 0.11 |
Gm49944 |
predicted gene, 49944 |
3420 |
0.26 |
| chr13_78162894_78163098 | 0.10 |
3110006O06Rik |
RIKEN cDNA 3110006O06 gene |
8634 |
0.14 |
| chr7_140103499_140103668 | 0.09 |
Fuom |
fucose mutarotase |
1142 |
0.28 |
| chr7_120173945_120174376 | 0.09 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
302 |
0.86 |
| chr13_113892909_113893095 | 0.08 |
Arl15 |
ADP-ribosylation factor-like 15 |
98380 |
0.06 |
| chr19_59066158_59066318 | 0.08 |
Shtn1 |
shootin 1 |
9831 |
0.19 |
| chr4_135494101_135494456 | 0.08 |
Nipal3 |
NIPA-like domain containing 3 |
228 |
0.72 |
| chr8_46837939_46838484 | 0.08 |
Gm45481 |
predicted gene 45481 |
4299 |
0.21 |
| chr5_122293071_122293465 | 0.07 |
Pptc7 |
PTC7 protein phosphatase homolog |
2504 |
0.17 |
| chr10_63156936_63157252 | 0.07 |
Mypn |
myopalladin |
34410 |
0.1 |
| chr8_111033405_111033617 | 0.07 |
Aars |
alanyl-tRNA synthetase |
367 |
0.72 |
| chr9_66575232_66575707 | 0.07 |
Usp3 |
ubiquitin specific peptidase 3 |
569 |
0.74 |
| chr10_3562232_3562413 | 0.06 |
Iyd |
iodotyrosine deiodinase |
22034 |
0.25 |
| chr4_95659510_95659661 | 0.06 |
Fggy |
FGGY carbohydrate kinase domain containing |
22698 |
0.23 |
| chr11_83849884_83850450 | 0.05 |
Hnf1b |
HNF1 homeobox B |
104 |
0.9 |
| chr7_24904782_24904948 | 0.05 |
Arhgef1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
425 |
0.68 |
| chr6_145855716_145856207 | 0.05 |
Gm43909 |
predicted gene, 43909 |
7336 |
0.17 |
| chr14_79632502_79632873 | 0.05 |
Sugt1 |
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
8048 |
0.17 |
| chr2_28622596_28622770 | 0.04 |
Gfi1b |
growth factor independent 1B |
701 |
0.54 |
| chrX_101944862_101945072 | 0.04 |
Nhsl2 |
NHS-like 2 |
16830 |
0.16 |
| chr15_99973613_99973793 | 0.04 |
2310068J16Rik |
RIKEN cDNA 2310068J16 gene |
416 |
0.59 |
| chr13_21996611_21996762 | 0.03 |
Prss16 |
protease, serine 16 (thymus) |
8536 |
0.06 |
| chr5_135113666_135114097 | 0.03 |
Mlxipl |
MLX interacting protein-like |
6963 |
0.1 |
| chr5_112302729_112302906 | 0.03 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
1079 |
0.37 |
| chr7_119895431_119895670 | 0.03 |
Dcun1d3 |
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
7 |
0.82 |
| chr10_128548464_128548828 | 0.02 |
Rpl41 |
ribosomal protein L41 |
456 |
0.48 |
| chr4_149425774_149426125 | 0.02 |
Ube4b |
ubiquitination factor E4B |
36 |
0.96 |
| chr9_32223889_32224219 | 0.02 |
Arhgap32 |
Rho GTPase activating protein 32 |
195 |
0.95 |
| chr4_138185094_138185245 | 0.01 |
Eif4g3 |
eukaryotic translation initiation factor 4 gamma, 3 |
691 |
0.6 |
| chr13_55182429_55182623 | 0.01 |
Fgfr4 |
fibroblast growth factor receptor 4 |
26409 |
0.11 |
| chr5_147077099_147077250 | 0.01 |
Polr1d |
polymerase (RNA) I polypeptide D |
124 |
0.87 |
| chr15_55563893_55564060 | 0.01 |
Mrpl13 |
mitochondrial ribosomal protein L13 |
6228 |
0.21 |
| chr9_113115901_113116052 | 0.01 |
Gm36251 |
predicted gene, 36251 |
7053 |
0.29 |
| chr4_136311666_136311888 | 0.01 |
Hnrnpr |
heterogeneous nuclear ribonucleoprotein R |
92 |
0.96 |
| chr1_120849519_120849684 | 0.01 |
Gm29347 |
predicted gene 29347 |
7073 |
0.15 |
| chr18_21216297_21216448 | 0.01 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
83751 |
0.08 |
| chr1_187763468_187763822 | 0.00 |
AC121143.1 |
NADH dehydrogenase 2, mitochondrial (mt-Nd2) pseudogene |
50840 |
0.16 |
| chr13_63046735_63046900 | 0.00 |
Aopep |
aminopeptidase O |
21298 |
0.16 |
| chr18_61637893_61638063 | 0.00 |
Bvht |
braveheart long non-coding RNA |
1564 |
0.26 |
| chr10_126978694_126978988 | 0.00 |
Ctdsp2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
124 |
0.91 |
| chr12_70913520_70913679 | 0.00 |
Gm10457 |
predicted gene 10457 |
1340 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) regulation of iron ion transport(GO:0034756) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0043733 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |