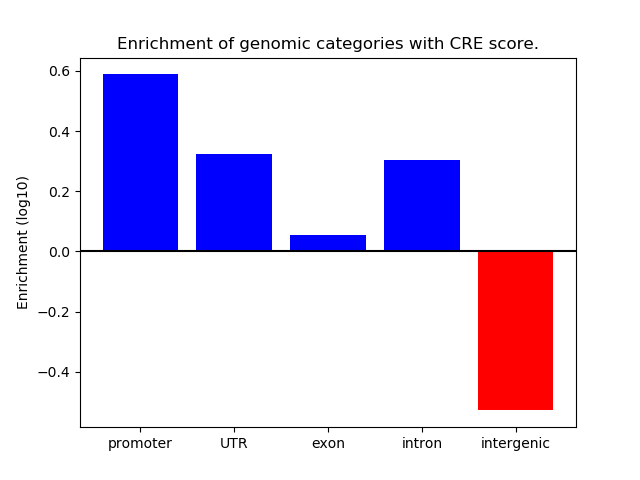
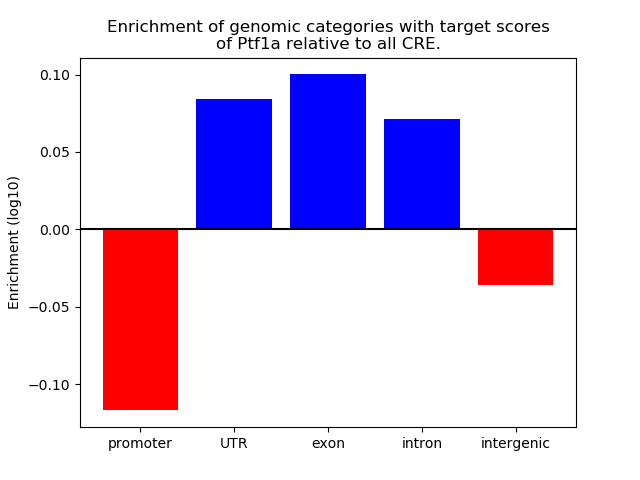
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
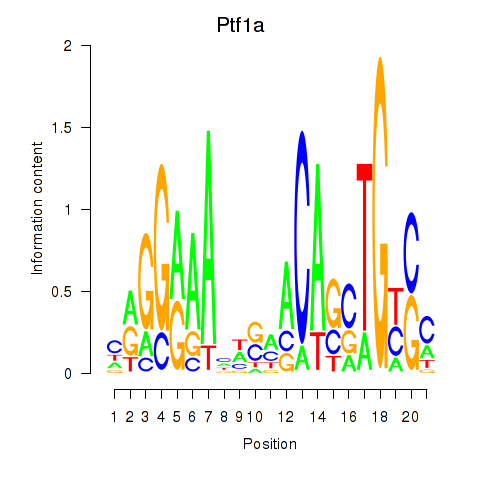
Results for Ptf1a
Z-value: 0.97
Transcription factors associated with Ptf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ptf1a
|
ENSMUSG00000026735.2 | pancreas specific transcription factor, 1a |
Activity of the Ptf1a motif across conditions
Conditions sorted by the z-value of the Ptf1a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
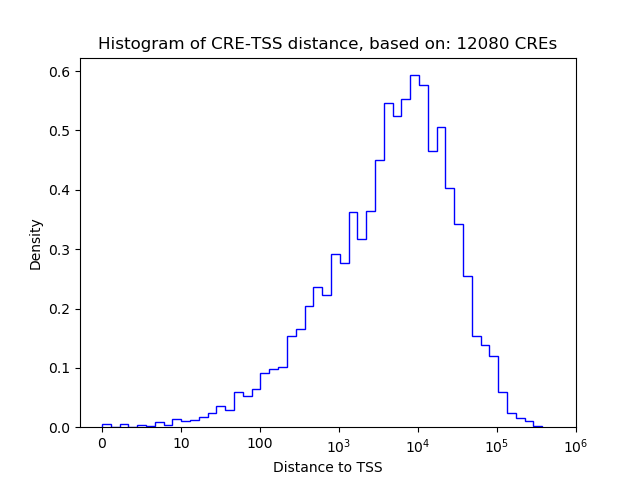
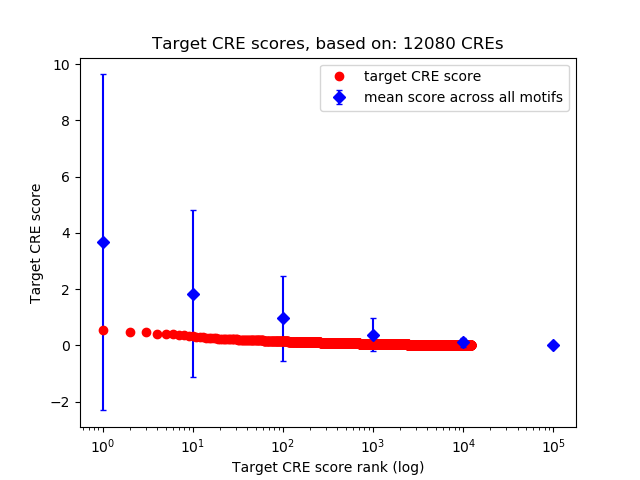
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_89140809_89141081 | 0.53 |
Pklr |
pyruvate kinase liver and red blood cell |
4322 |
0.08 |
| chr19_10599348_10599499 | 0.49 |
Tkfc |
triokinase, FMN cyclase |
4835 |
0.1 |
| chr4_141955631_141955810 | 0.48 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
1526 |
0.3 |
| chr2_134708873_134709037 | 0.39 |
Gm14037 |
predicted gene 14037 |
22152 |
0.2 |
| chr13_62879517_62879679 | 0.39 |
Fbp1 |
fructose bisphosphatase 1 |
6840 |
0.14 |
| chr3_18143898_18144049 | 0.39 |
Gm23686 |
predicted gene, 23686 |
33652 |
0.17 |
| chr11_16826830_16827159 | 0.35 |
Egfros |
epidermal growth factor receptor, opposite strand |
3708 |
0.27 |
| chr5_135728456_135728639 | 0.35 |
Por |
P450 (cytochrome) oxidoreductase |
2819 |
0.15 |
| chr4_137307447_137307614 | 0.32 |
Gm25772 |
predicted gene, 25772 |
18029 |
0.13 |
| chr2_21211497_21211673 | 0.31 |
Thnsl1 |
threonine synthase-like 1 (bacterial) |
5798 |
0.17 |
| chr4_45100494_45100645 | 0.31 |
Tomm5 |
translocase of outer mitochondrial membrane 5 |
7469 |
0.15 |
| chr11_50131029_50131991 | 0.30 |
Tbc1d9b |
TBC1 domain family, member 9B |
108 |
0.94 |
| chr11_98766772_98767040 | 0.29 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3520 |
0.12 |
| chr18_65968667_65968818 | 0.27 |
Cplx4 |
complexin 4 |
1060 |
0.37 |
| chr11_70389533_70389684 | 0.26 |
Pelp1 |
proline, glutamic acid and leucine rich protein 1 |
20420 |
0.08 |
| chr11_16856758_16857153 | 0.26 |
Egfr |
epidermal growth factor receptor |
21195 |
0.18 |
| chr11_116206084_116206272 | 0.25 |
Ten1 |
TEN1 telomerase capping complex subunit |
4756 |
0.1 |
| chr11_16836874_16837160 | 0.25 |
Egfros |
epidermal growth factor receptor, opposite strand |
6315 |
0.23 |
| chr6_51723156_51723321 | 0.24 |
Gm38811 |
predicted gene, 38811 |
12157 |
0.25 |
| chr9_106234370_106234560 | 0.24 |
Alas1 |
aminolevulinic acid synthase 1 |
2619 |
0.15 |
| chr15_84133579_84133730 | 0.23 |
Gm33432 |
predicted gene, 33432 |
6350 |
0.11 |
| chr7_34718747_34718898 | 0.23 |
Chst8 |
carbohydrate sulfotransferase 8 |
21372 |
0.16 |
| chr13_48883902_48884053 | 0.22 |
Phf2 |
PHD finger protein 2 |
12858 |
0.19 |
| chr5_148909547_148909715 | 0.22 |
Gm43600 |
predicted gene 43600 |
17057 |
0.11 |
| chr11_60201760_60201955 | 0.22 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
358 |
0.51 |
| chr4_95207881_95208040 | 0.22 |
Gm12708 |
predicted gene 12708 |
14598 |
0.2 |
| chr11_120816318_120816478 | 0.21 |
Fasn |
fatty acid synthase |
943 |
0.36 |
| chr5_65349117_65349297 | 0.21 |
Klb |
klotho beta |
799 |
0.5 |
| chr11_98768301_98768532 | 0.21 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2010 |
0.18 |
| chr5_90639561_90639792 | 0.21 |
Rassf6 |
Ras association (RalGDS/AF-6) domain family member 6 |
811 |
0.61 |
| chr3_146605344_146605733 | 0.21 |
Uox |
urate oxidase |
4447 |
0.15 |
| chr19_44790216_44790367 | 0.20 |
Gm20395 |
predicted gene 20395 |
5779 |
0.15 |
| chr7_97569323_97569597 | 0.20 |
Aamdc |
adipogenesis associated Mth938 domain containing |
6193 |
0.13 |
| chr4_154705761_154705932 | 0.20 |
Actrt2 |
actin-related protein T2 |
37979 |
0.12 |
| chr9_107660530_107661377 | 0.20 |
Slc38a3 |
solute carrier family 38, member 3 |
1425 |
0.2 |
| chrX_42602450_42602620 | 0.20 |
Sh2d1a |
SH2 domain containing 1A |
75950 |
0.1 |
| chr6_91599085_91599288 | 0.19 |
Gm45218 |
predicted gene 45218 |
46 |
0.97 |
| chr10_8817897_8818050 | 0.19 |
Gm25410 |
predicted gene, 25410 |
6174 |
0.21 |
| chr7_101106567_101106718 | 0.19 |
Fchsd2 |
FCH and double SH3 domains 2 |
2091 |
0.29 |
| chrY_90798569_90798761 | 0.19 |
Gm47283 |
predicted gene, 47283 |
8214 |
0.18 |
| chr2_147811436_147811589 | 0.19 |
Gm25516 |
predicted gene, 25516 |
63366 |
0.12 |
| chr6_66957937_66958096 | 0.18 |
Gm36816 |
predicted gene, 36816 |
50397 |
0.08 |
| chr19_26716546_26716697 | 0.18 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
506 |
0.84 |
| chr7_19756083_19756234 | 0.18 |
Bcam |
basal cell adhesion molecule |
793 |
0.37 |
| chr8_84723608_84723805 | 0.18 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
699 |
0.52 |
| chr11_120813788_120814608 | 0.18 |
Fasn |
fatty acid synthase |
1257 |
0.26 |
| chr4_35916085_35916245 | 0.18 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
70961 |
0.13 |
| chr4_102850618_102850787 | 0.18 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
19862 |
0.23 |
| chr6_108519143_108519315 | 0.18 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
497 |
0.75 |
| chr10_87937503_87937727 | 0.18 |
Tyms-ps |
thymidylate synthase, pseudogene |
29232 |
0.15 |
| chr5_139358617_139359046 | 0.18 |
Cyp2w1 |
cytochrome P450, family 2, subfamily w, polypeptide 1 |
4255 |
0.13 |
| chr11_120819185_120819682 | 0.18 |
Fasn |
fatty acid synthase |
3978 |
0.11 |
| chr11_53307027_53307178 | 0.17 |
Hspa4 |
heat shock protein 4 |
6645 |
0.15 |
| chr9_74892742_74892933 | 0.17 |
Onecut1 |
one cut domain, family member 1 |
26353 |
0.14 |
| chr4_49553429_49553778 | 0.17 |
Aldob |
aldolase B, fructose-bisphosphate |
4057 |
0.17 |
| chr5_113141351_113141509 | 0.17 |
4930557B06Rik |
RIKEN cDNA 4930557B06 gene |
1019 |
0.36 |
| chr8_77310146_77310297 | 0.17 |
4933421D24Rik |
RIKEN cDNA 4933421D24 gene |
4688 |
0.2 |
| chr3_51252801_51253044 | 0.17 |
Elf2 |
E74-like factor 2 |
7319 |
0.13 |
| chr6_49095060_49095211 | 0.17 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
924 |
0.43 |
| chr19_23160573_23160812 | 0.17 |
Mir1192 |
microRNA 1192 |
11261 |
0.16 |
| chr10_19443329_19443501 | 0.17 |
Gm33104 |
predicted gene, 33104 |
43300 |
0.15 |
| chr12_104343910_104344587 | 0.17 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
5762 |
0.12 |
| chr12_16610798_16610973 | 0.17 |
Lpin1 |
lipin 1 |
81 |
0.98 |
| chr18_9743359_9743510 | 0.17 |
Gm7497 |
predicted gene 7497 |
5298 |
0.13 |
| chr15_83202527_83202771 | 0.17 |
7530414M10Rik |
RIKEN cDNA 7530414M10 gene |
26204 |
0.1 |
| chr15_36270573_36270724 | 0.16 |
Rnf19a |
ring finger protein 19A |
12450 |
0.11 |
| chr4_102605309_102605498 | 0.16 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
15560 |
0.27 |
| chr4_111200292_111200482 | 0.16 |
Agbl4 |
ATP/GTP binding protein-like 4 |
81476 |
0.1 |
| chr16_12233397_12233554 | 0.16 |
Gm19571 |
predicted gene, 19571 |
148027 |
0.04 |
| chr6_90771679_90771830 | 0.16 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
7613 |
0.18 |
| chr4_43558445_43558602 | 0.16 |
Tln1 |
talin 1 |
63 |
0.93 |
| chr3_36533351_36533502 | 0.16 |
Gm11549 |
predicted gene 11549 |
1042 |
0.38 |
| chr8_61585782_61586047 | 0.16 |
Palld |
palladin, cytoskeletal associated protein |
5225 |
0.3 |
| chr7_132724267_132724477 | 0.16 |
Fam53b |
family with sequence similarity 53, member B |
52544 |
0.11 |
| chr6_108667548_108667923 | 0.16 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
4689 |
0.18 |
| chr16_26669940_26670091 | 0.16 |
Il1rap |
interleukin 1 receptor accessory protein |
45859 |
0.18 |
| chr13_92277409_92277573 | 0.16 |
Msh3 |
mutS homolog 3 |
11419 |
0.21 |
| chr1_100386126_100386277 | 0.16 |
Gm29667 |
predicted gene 29667 |
103410 |
0.07 |
| chr9_121677553_121677957 | 0.16 |
Vipr1 |
vasoactive intestinal peptide receptor 1 |
9042 |
0.11 |
| chr8_124267313_124267488 | 0.15 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
27703 |
0.14 |
| chr5_114246590_114246772 | 0.15 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
1652 |
0.29 |
| chr17_81175582_81175959 | 0.15 |
Gm50042 |
predicted gene, 50042 |
70968 |
0.11 |
| chr3_67430526_67430935 | 0.15 |
Gfm1 |
G elongation factor, mitochondrial 1 |
614 |
0.7 |
| chr11_120803051_120803202 | 0.15 |
Fasn |
fatty acid synthase |
5479 |
0.09 |
| chr10_4589602_4589901 | 0.15 |
Gm3266 |
predicted gene 3266 |
19528 |
0.18 |
| chr16_35315588_35315746 | 0.15 |
Gm49706 |
predicted gene, 49706 |
7399 |
0.16 |
| chr7_38162494_38162645 | 0.15 |
Gm22203 |
predicted gene, 22203 |
14102 |
0.13 |
| chr18_14962680_14963008 | 0.15 |
4933424G05Rik |
RIKEN cDNA 4933424G05 gene |
26151 |
0.15 |
| chr18_84376572_84376752 | 0.15 |
Gm37216 |
predicted gene, 37216 |
516 |
0.84 |
| chr8_40995175_40995326 | 0.15 |
Mtus1 |
mitochondrial tumor suppressor 1 |
4955 |
0.19 |
| chr12_103893329_103893685 | 0.15 |
B430119L08Rik |
RIKEN cDNA B430119L08 gene |
2390 |
0.16 |
| chr10_75940185_75940352 | 0.15 |
Gm867 |
predicted gene 867 |
343 |
0.7 |
| chr2_34777844_34777997 | 0.15 |
Hspa5 |
heat shock protein 5 |
3073 |
0.17 |
| chr5_143770909_143771060 | 0.15 |
Gm42503 |
predicted gene 42503 |
944 |
0.5 |
| chr5_146191113_146191264 | 0.15 |
B230303O12Rik |
RIKEN cDNA B230303O12 gene |
3621 |
0.12 |
| chr19_28347532_28347702 | 0.15 |
Glis3 |
GLIS family zinc finger 3 |
21611 |
0.26 |
| chr3_79563130_79563296 | 0.14 |
Fnip2 |
folliculin interacting protein 2 |
4466 |
0.13 |
| chr13_24360335_24360523 | 0.14 |
Gm11342 |
predicted gene 11342 |
15521 |
0.12 |
| chr2_93132224_93132381 | 0.14 |
Gm13802 |
predicted gene 13802 |
34058 |
0.16 |
| chr14_17120556_17120707 | 0.14 |
Gm48312 |
predicted gene, 48312 |
872 |
0.68 |
| chr8_111739071_111739223 | 0.14 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
4662 |
0.21 |
| chr2_127367630_127367825 | 0.14 |
Adra2b |
adrenergic receptor, alpha 2b |
4441 |
0.16 |
| chr9_55232715_55232911 | 0.14 |
Nrg4 |
neuregulin 4 |
9648 |
0.17 |
| chr14_105803498_105803761 | 0.14 |
Gm22406 |
predicted gene, 22406 |
13279 |
0.23 |
| chr12_70212633_70212998 | 0.14 |
Pygl |
liver glycogen phosphorylase |
5467 |
0.16 |
| chr9_74523576_74523891 | 0.14 |
Gm28622 |
predicted gene 28622 |
37645 |
0.17 |
| chr17_87288069_87288478 | 0.14 |
Ttc7 |
tetratricopeptide repeat domain 7 |
4538 |
0.16 |
| chr7_81599959_81600110 | 0.14 |
Gm45698 |
predicted gene 45698 |
9140 |
0.11 |
| chr8_56335584_56335764 | 0.14 |
Gm45540 |
predicted gene 45540 |
24370 |
0.2 |
| chr16_70320670_70320837 | 0.14 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
6619 |
0.26 |
| chr7_45710445_45711101 | 0.14 |
Sphk2 |
sphingosine kinase 2 |
2670 |
0.09 |
| chr14_69408066_69408257 | 0.14 |
Gm38378 |
predicted gene, 38378 |
10390 |
0.11 |
| chr2_58773911_58774203 | 0.14 |
Upp2 |
uridine phosphorylase 2 |
8732 |
0.21 |
| chr14_45453394_45453727 | 0.14 |
Gm34250 |
predicted gene, 34250 |
6487 |
0.13 |
| chr7_24919736_24919914 | 0.14 |
Arhgef1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
623 |
0.53 |
| chr8_24435886_24436037 | 0.13 |
Tcim |
transcriptional and immune response regulator |
3023 |
0.2 |
| chr10_80283389_80283610 | 0.13 |
Gm15122 |
predicted gene 15122 |
493 |
0.41 |
| chr1_181035644_181035840 | 0.13 |
Gm37086 |
predicted gene, 37086 |
9464 |
0.09 |
| chr3_28712599_28712834 | 0.13 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
8801 |
0.17 |
| chr3_89147086_89147561 | 0.13 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
3891 |
0.08 |
| chr18_61357667_61357827 | 0.13 |
Gm25301 |
predicted gene, 25301 |
40066 |
0.1 |
| chr1_67236092_67236243 | 0.13 |
Gm15668 |
predicted gene 15668 |
13033 |
0.22 |
| chr5_34987481_34987632 | 0.13 |
Rgs12 |
regulator of G-protein signaling 12 |
1572 |
0.35 |
| chr13_96567943_96568116 | 0.13 |
Gm46436 |
predicted gene, 46436 |
8601 |
0.12 |
| chr14_33472162_33472313 | 0.13 |
Frmpd2 |
FERM and PDZ domain containing 2 |
429 |
0.82 |
| chr14_69626340_69626524 | 0.13 |
Gm37513 |
predicted gene, 37513 |
10386 |
0.11 |
| chr17_57207104_57207255 | 0.13 |
Mir6978 |
microRNA 6978 |
10049 |
0.1 |
| chr10_4621590_4621757 | 0.13 |
Esr1 |
estrogen receptor 1 (alpha) |
9652 |
0.24 |
| chr15_7163262_7163413 | 0.13 |
Lifr |
LIF receptor alpha |
8984 |
0.27 |
| chr11_117595990_117596164 | 0.13 |
2900041M22Rik |
RIKEN cDNA 2900041M22 gene |
15170 |
0.18 |
| chr15_59476623_59476975 | 0.13 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
102512 |
0.06 |
| chr19_39507413_39507564 | 0.13 |
Cyp2c39 |
cytochrome P450, family 2, subfamily c, polypeptide 39 |
3374 |
0.31 |
| chr13_55211103_55211935 | 0.13 |
Nsd1 |
nuclear receptor-binding SET-domain protein 1 |
22 |
0.97 |
| chr7_97423614_97423975 | 0.13 |
Thrsp |
thyroid hormone responsive |
6064 |
0.14 |
| chr4_101428291_101428631 | 0.13 |
Ak4 |
adenylate kinase 4 |
7964 |
0.16 |
| chr8_76535740_76535905 | 0.13 |
Gm27355 |
predicted gene, 27355 |
83460 |
0.1 |
| chr7_26835515_26835776 | 0.13 |
Cyp2a5 |
cytochrome P450, family 2, subfamily a, polypeptide 5 |
281 |
0.9 |
| chr11_6021664_6021840 | 0.13 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
21304 |
0.15 |
| chr4_49535372_49535819 | 0.13 |
Aldob |
aldolase B, fructose-bisphosphate |
3271 |
0.17 |
| chr6_28524650_28524824 | 0.13 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
37080 |
0.11 |
| chr15_58980059_58980524 | 0.13 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
3845 |
0.19 |
| chr1_153722020_153722196 | 0.13 |
5530400K19Rik |
RIKEN cDNA 5530400K19 gene |
7454 |
0.11 |
| chr6_55384309_55384460 | 0.13 |
Ghrhr |
growth hormone releasing hormone receptor |
8019 |
0.17 |
| chr19_10089080_10089259 | 0.13 |
Fads2 |
fatty acid desaturase 2 |
453 |
0.77 |
| chr17_36920402_36920584 | 0.13 |
1700031A10Rik |
RIKEN cDNA 1700031A10 gene |
3299 |
0.1 |
| chr13_110845837_110846005 | 0.13 |
Gm48534 |
predicted gene, 48534 |
985 |
0.49 |
| chr17_26113500_26114085 | 0.13 |
Tmem8 |
transmembrane protein 8 |
449 |
0.63 |
| chr6_85343141_85343317 | 0.12 |
Rab11fip5 |
RAB11 family interacting protein 5 (class I) |
1592 |
0.3 |
| chr8_84723357_84723534 | 0.12 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
438 |
0.7 |
| chr6_119366700_119366851 | 0.12 |
Adipor2 |
adiponectin receptor 2 |
21911 |
0.17 |
| chr11_60201956_60202762 | 0.12 |
Mir6922 |
microRNA 6922 |
32 |
0.63 |
| chr17_78587278_78587447 | 0.12 |
Vit |
vitrin |
13417 |
0.2 |
| chr17_5926825_5927009 | 0.12 |
Gm8376 |
predicted gene 8376 |
12170 |
0.17 |
| chr17_75539211_75539426 | 0.12 |
Fam98a |
family with sequence similarity 98, member A |
1254 |
0.57 |
| chr9_106233703_106233854 | 0.12 |
Alas1 |
aminolevulinic acid synthase 1 |
3306 |
0.13 |
| chr19_32210467_32210618 | 0.12 |
Sgms1 |
sphingomyelin synthase 1 |
471 |
0.75 |
| chr1_174906254_174906499 | 0.12 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
15443 |
0.29 |
| chr5_113136409_113136645 | 0.12 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
1459 |
0.26 |
| chr10_89471762_89471916 | 0.12 |
Gas2l3 |
growth arrest-specific 2 like 3 |
27872 |
0.18 |
| chr7_119634973_119635124 | 0.12 |
Gm45792 |
predicted gene 45792 |
3990 |
0.13 |
| chr3_87801773_87801942 | 0.12 |
Insrr |
insulin receptor-related receptor |
86 |
0.95 |
| chr1_192697997_192698159 | 0.12 |
Hhat |
hedgehog acyltransferase |
10372 |
0.22 |
| chr11_119856806_119856957 | 0.12 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
563 |
0.72 |
| chr15_102103646_102103840 | 0.12 |
Tns2 |
tensin 2 |
682 |
0.55 |
| chr11_77781126_77781437 | 0.12 |
Myo18a |
myosin XVIIIA |
3965 |
0.17 |
| chr5_125517764_125517915 | 0.12 |
Aacs |
acetoacetyl-CoA synthetase |
2596 |
0.23 |
| chr11_120815283_120815743 | 0.12 |
Fasn |
fatty acid synthase |
58 |
0.94 |
| chr12_103302328_103302479 | 0.12 |
Fam181a |
family with sequence similarity 181, member A |
8572 |
0.13 |
| chr2_131321117_131321286 | 0.12 |
Snrpert |
small nuclear ribonucleoprotein E, pseudogene |
1165 |
0.36 |
| chr9_105502550_105502701 | 0.12 |
Atp2c1 |
ATPase, Ca++-sequestering |
7130 |
0.18 |
| chr7_112688480_112688662 | 0.12 |
Tead1 |
TEA domain family member 1 |
3127 |
0.21 |
| chr3_52290025_52290176 | 0.12 |
Gm38034 |
predicted gene, 38034 |
7859 |
0.15 |
| chr19_40153747_40153898 | 0.12 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr4_133741821_133742020 | 0.12 |
Arid1a |
AT rich interactive domain 1A (SWI-like) |
11691 |
0.14 |
| chr11_86584132_86584307 | 0.12 |
Mir21a |
microRNA 21a |
61 |
0.96 |
| chr5_100393872_100394036 | 0.12 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
1341 |
0.38 |
| chr4_53204101_53204252 | 0.12 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
13385 |
0.18 |
| chr2_3760255_3760454 | 0.11 |
Gm13185 |
predicted gene 13185 |
5177 |
0.2 |
| chr11_98766479_98766640 | 0.11 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3867 |
0.12 |
| chr2_39160826_39161015 | 0.11 |
Gm16523 |
predicted gene, 16523 |
1069 |
0.39 |
| chr11_100524948_100525284 | 0.11 |
Acly |
ATP citrate lyase |
1518 |
0.24 |
| chr5_90465453_90465660 | 0.11 |
Alb |
albumin |
2856 |
0.22 |
| chr4_8220995_8221186 | 0.11 |
Gm25355 |
predicted gene, 25355 |
14492 |
0.2 |
| chr10_4599676_4599928 | 0.11 |
Esr1 |
estrogen receptor 1 (alpha) |
11791 |
0.21 |
| chr4_46868313_46868486 | 0.11 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
8497 |
0.27 |
| chr4_47258068_47258263 | 0.11 |
Col15a1 |
collagen, type XV, alpha 1 |
370 |
0.89 |
| chr6_51678830_51679023 | 0.11 |
Gm38811 |
predicted gene, 38811 |
32155 |
0.18 |
| chr6_31122323_31122530 | 0.11 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
11506 |
0.12 |
| chr3_97653476_97653627 | 0.11 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
4642 |
0.15 |
| chr3_146607555_146607962 | 0.11 |
Uox |
urate oxidase |
2227 |
0.22 |
| chr4_72766654_72766805 | 0.11 |
Aldoart1 |
aldolase 1 A, retrogene 1 |
85903 |
0.09 |
| chr2_73591851_73592008 | 0.11 |
Chn1os3 |
chimerin 1, opposite strand 3 |
4597 |
0.16 |
| chr3_27625698_27625849 | 0.11 |
Mir3092 |
microRNA 3092 |
40778 |
0.17 |
| chr8_91332717_91332888 | 0.11 |
Fto |
fat mass and obesity associated |
19188 |
0.13 |
| chr11_90383773_90384100 | 0.11 |
Hlf |
hepatic leukemia factor |
2001 |
0.43 |
| chr6_128662913_128663074 | 0.11 |
Clec2h |
C-type lectin domain family 2, member h |
602 |
0.46 |
| chr13_64187132_64187283 | 0.11 |
Habp4 |
hyaluronic acid binding protein 4 |
2704 |
0.17 |
| chr3_24779536_24779704 | 0.11 |
Naaladl2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
4002 |
0.37 |
| chr15_58944330_58944500 | 0.11 |
Mtss1 |
MTSS I-BAR domain containing 1 |
9165 |
0.13 |
| chr18_67447402_67447553 | 0.11 |
Afg3l2 |
AFG3-like AAA ATPase 2 |
1630 |
0.3 |
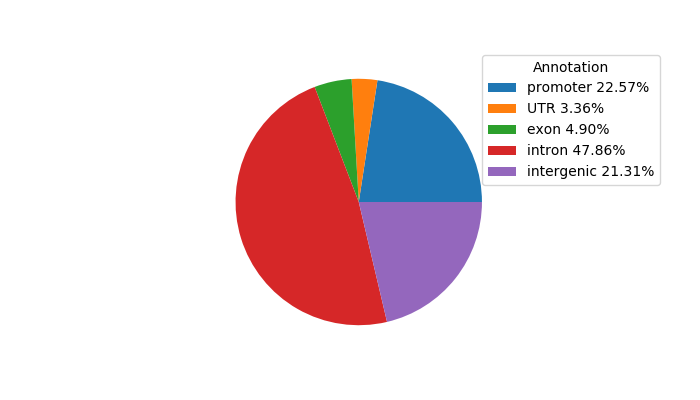
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.0 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.0 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.0 | GO:1904751 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.0 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.0 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.4 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0035276 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |