Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
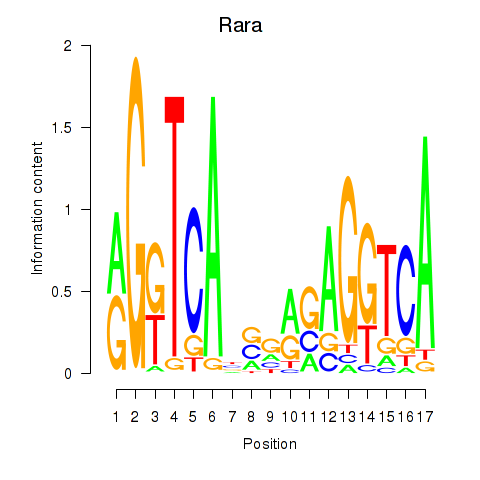
Results for Rara
Z-value: 1.16
Transcription factors associated with Rara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rara
|
ENSMUSG00000037992.10 | retinoic acid receptor, alpha |
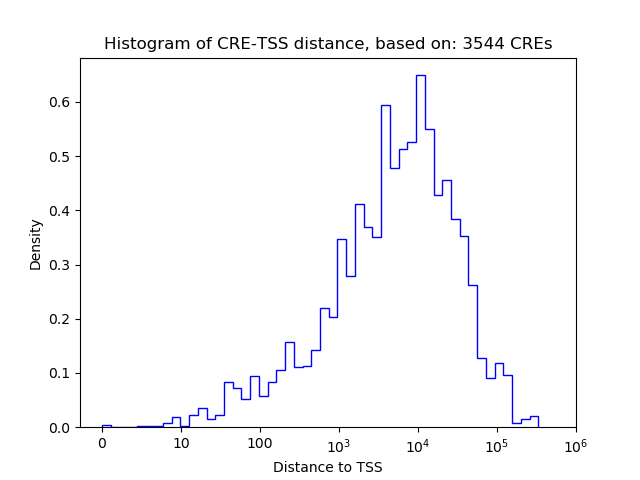
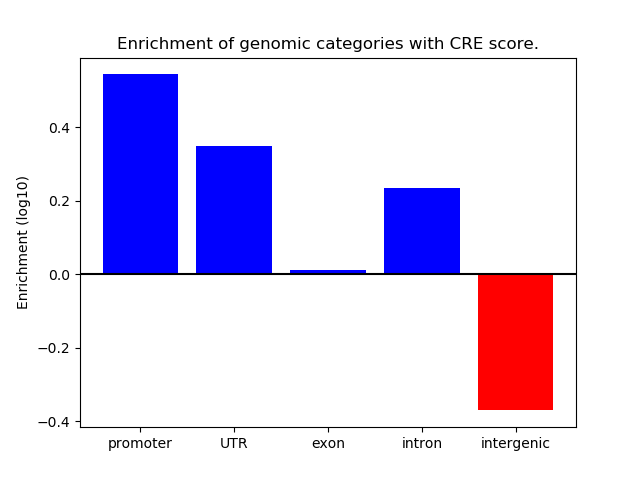
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_98971402_98971560 | Rara | 11069 | 0.098839 | -0.95 | 4.0e-03 | Click! |
| chr11_98941866_98942032 | Rara | 2237 | 0.175688 | 0.88 | 2.1e-02 | Click! |
| chr11_98942156_98942339 | Rara | 2535 | 0.159794 | 0.88 | 2.1e-02 | Click! |
| chr11_98937139_98937314 | Rara | 472 | 0.689095 | 0.86 | 2.8e-02 | Click! |
| chr11_98964767_98965121 | Rara | 4532 | 0.119697 | 0.86 | 2.9e-02 | Click! |
Activity of the Rara motif across conditions
Conditions sorted by the z-value of the Rara motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
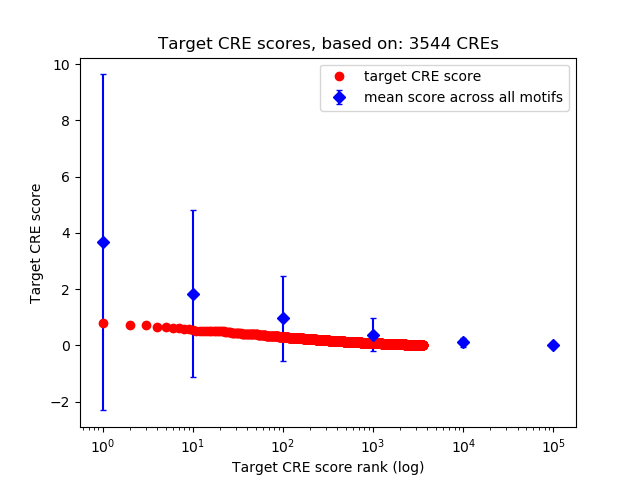
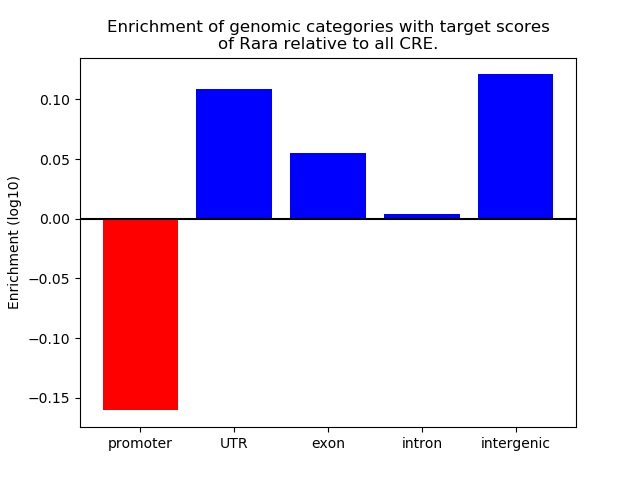
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_115831765_115832080 | 0.80 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
124128 |
0.06 |
| chr3_145834049_145834216 | 0.74 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
23653 |
0.18 |
| chr6_126404787_126404969 | 0.72 |
Gm44431 |
predicted gene, 44431 |
68011 |
0.1 |
| chr7_140948066_140948469 | 0.66 |
Ifitm5 |
interferon induced transmembrane protein 5 |
2024 |
0.12 |
| chr11_53872113_53872322 | 0.66 |
Slc22a5 |
solute carrier family 22 (organic cation transporter), member 5 |
540 |
0.66 |
| chr4_138440154_138440340 | 0.62 |
Mul1 |
mitochondrial ubiquitin ligase activator of NFKB 1 |
5524 |
0.14 |
| chr5_135738165_135738332 | 0.61 |
Tmem120a |
transmembrane protein 120A |
1012 |
0.35 |
| chr1_191663564_191663926 | 0.58 |
Lpgat1 |
lysophosphatidylglycerol acyltransferase 1 |
54089 |
0.09 |
| chr8_4193425_4193618 | 0.57 |
Evi5l |
ecotropic viral integration site 5 like |
2229 |
0.17 |
| chr5_66200133_66200302 | 0.56 |
Gm15794 |
predicted gene 15794 |
1109 |
0.34 |
| chr8_84131316_84131478 | 0.53 |
Podnl1 |
podocan-like 1 |
3579 |
0.09 |
| chr6_82753132_82753283 | 0.52 |
Gm17034 |
predicted gene 17034 |
11075 |
0.14 |
| chr4_6539560_6539735 | 0.52 |
Gm11801 |
predicted gene 11801 |
86 |
0.98 |
| chr1_58475799_58475950 | 0.52 |
Orc2 |
origin recognition complex, subunit 2 |
5575 |
0.13 |
| chr7_126260333_126260533 | 0.51 |
Sbk1 |
SH3-binding kinase 1 |
11571 |
0.11 |
| chr2_12197838_12198004 | 0.50 |
Itga8 |
integrin alpha 8 |
4609 |
0.27 |
| chr18_36527710_36528079 | 0.50 |
Slc4a9 |
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
253 |
0.88 |
| chr4_131918798_131918960 | 0.50 |
Gm12992 |
predicted gene 12992 |
1150 |
0.29 |
| chr1_170501557_170501869 | 0.50 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
46799 |
0.15 |
| chr5_134571935_134572111 | 0.50 |
Mir7228 |
microRNA 7228 |
53 |
0.95 |
| chr11_115303297_115303464 | 0.49 |
Otop2 |
otopetrin 2 |
3783 |
0.11 |
| chr9_98429903_98430078 | 0.49 |
Rbp1 |
retinol binding protein 1, cellular |
7029 |
0.21 |
| chr6_94998536_94998699 | 0.48 |
4930511E03Rik |
RIKEN cDNA 4930511E03 gene |
54783 |
0.13 |
| chr5_103783262_103783453 | 0.48 |
Aff1 |
AF4/FMR2 family, member 1 |
1002 |
0.59 |
| chr15_97059709_97059903 | 0.47 |
Slc38a4 |
solute carrier family 38, member 4 |
3850 |
0.33 |
| chr15_16566506_16566694 | 0.46 |
Gm2611 |
predicted gene 2611 |
33962 |
0.19 |
| chr15_10335815_10335966 | 0.45 |
Gm21973 |
predicted gene 21973 |
10510 |
0.19 |
| chr13_97453178_97453337 | 0.45 |
Gm8756 |
predicted gene 8756 |
5820 |
0.18 |
| chr1_140183261_140183634 | 0.44 |
Cfh |
complement component factor h |
36 |
0.98 |
| chr7_19965081_19965263 | 0.44 |
Ceacam20 |
carcinoembryonic antigen-related cell adhesion molecule 20 |
240 |
0.83 |
| chr15_79434871_79435022 | 0.44 |
Csnk1e |
casein kinase 1, epsilon |
1820 |
0.21 |
| chr11_120500689_120500856 | 0.43 |
Slc25a10 |
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
3047 |
0.09 |
| chr8_10531058_10531225 | 0.43 |
5330413D20Rik |
RIKEN cDNA 5330413D20 gene |
145772 |
0.04 |
| chr7_19865343_19865508 | 0.43 |
Ceacam16 |
carcinoembryonic antigen-related cell adhesion molecule 16 |
4126 |
0.08 |
| chr11_70585533_70585714 | 0.41 |
Mink1 |
misshapen-like kinase 1 (zebrafish) |
13365 |
0.07 |
| chr4_118366670_118366829 | 0.41 |
Szt2 |
SZT2 subunit of KICSTOR complex |
1683 |
0.28 |
| chrX_11574219_11574397 | 0.41 |
Gm14515 |
predicted gene 14515 |
27046 |
0.22 |
| chr13_52060383_52060534 | 0.41 |
Gm48190 |
predicted gene, 48190 |
36184 |
0.14 |
| chr9_44579542_44579697 | 0.41 |
Gm47230 |
predicted gene, 47230 |
8 |
0.94 |
| chr6_128481760_128482446 | 0.40 |
Pzp |
PZP, alpha-2-macroglobulin like |
5752 |
0.09 |
| chr7_68692846_68692997 | 0.40 |
Gm44692 |
predicted gene 44692 |
33546 |
0.17 |
| chr15_10338082_10338258 | 0.40 |
Gm21973 |
predicted gene 21973 |
12790 |
0.18 |
| chr14_65299395_65299546 | 0.40 |
Gm48433 |
predicted gene, 48433 |
26861 |
0.13 |
| chr11_120215894_120216125 | 0.39 |
Gm11770 |
predicted gene 11770 |
984 |
0.37 |
| chr2_158783086_158783239 | 0.39 |
Fam83d |
family with sequence similarity 83, member D |
1754 |
0.34 |
| chr3_118478170_118478340 | 0.39 |
Gm26871 |
predicted gene, 26871 |
20596 |
0.14 |
| chr5_64840100_64840327 | 0.39 |
Klf3 |
Kruppel-like factor 3 (basic) |
13404 |
0.15 |
| chr8_91706641_91706804 | 0.39 |
Gm36325 |
predicted gene, 36325 |
7691 |
0.14 |
| chr11_86917162_86917343 | 0.39 |
Ypel2 |
yippee like 2 |
54772 |
0.11 |
| chr18_20989972_20990130 | 0.39 |
Rnf138 |
ring finger protein 138 |
11290 |
0.2 |
| chr8_79498769_79498920 | 0.39 |
Gm8077 |
predicted gene 8077 |
666 |
0.57 |
| chr8_85064057_85064246 | 0.39 |
Fbxw9 |
F-box and WD-40 domain protein 9 |
444 |
0.52 |
| chr6_115730698_115730849 | 0.38 |
Tmem40 |
transmembrane protein 40 |
4178 |
0.13 |
| chr19_42891636_42891940 | 0.38 |
Hps1 |
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
111810 |
0.06 |
| chr2_153493974_153494180 | 0.38 |
4930404H24Rik |
RIKEN cDNA 4930404H24 gene |
1287 |
0.36 |
| chr19_8567498_8567799 | 0.38 |
Gm6425 |
predicted pseudogene 6425 |
21548 |
0.11 |
| chr8_119419822_119419973 | 0.37 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
14227 |
0.14 |
| chr1_62821658_62821837 | 0.37 |
Gm4208 |
predicted gene 4208 |
10623 |
0.19 |
| chr10_105404051_105404220 | 0.37 |
Gm48203 |
predicted gene, 48203 |
9849 |
0.19 |
| chr2_132740163_132740333 | 0.37 |
Gm22245 |
predicted gene, 22245 |
23142 |
0.11 |
| chr5_66458954_66459109 | 0.36 |
Gm43794 |
predicted gene 43794 |
14608 |
0.15 |
| chr15_78925213_78925506 | 0.36 |
Lgals1 |
lectin, galactose binding, soluble 1 |
1366 |
0.2 |
| chr10_107383971_107384142 | 0.35 |
Gm38117 |
predicted gene, 38117 |
53303 |
0.13 |
| chr9_96379613_96379768 | 0.35 |
Atp1b3 |
ATPase, Na+/K+ transporting, beta 3 polypeptide |
15248 |
0.17 |
| chr16_34965483_34965646 | 0.35 |
E130310I04Rik |
RIKEN cDNA E130310I04 gene |
6570 |
0.17 |
| chr17_14570462_14570613 | 0.35 |
Thbs2 |
thrombospondin 2 |
100329 |
0.06 |
| chr10_81102537_81102688 | 0.35 |
Mir5615-2 |
microRNA 5615-2 |
2004 |
0.12 |
| chr4_139978730_139978900 | 0.34 |
Klhdc7a |
kelch domain containing 7A |
10789 |
0.16 |
| chr3_88339093_88339486 | 0.34 |
Smg5 |
Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) |
1431 |
0.18 |
| chr11_117203569_117203863 | 0.34 |
Septin9 |
septin 9 |
4055 |
0.2 |
| chr11_82858816_82858975 | 0.34 |
Rffl |
ring finger and FYVE like domain containing protein |
11849 |
0.1 |
| chr5_63969602_63969775 | 0.34 |
Rell1 |
RELT-like 1 |
791 |
0.59 |
| chr10_8912248_8912575 | 0.34 |
Gm48727 |
predicted gene, 48727 |
8430 |
0.16 |
| chr11_113725344_113725520 | 0.34 |
Cdc42ep4 |
CDC42 effector protein (Rho GTPase binding) 4 |
13890 |
0.12 |
| chr7_29251255_29251571 | 0.34 |
2200002D01Rik |
RIKEN cDNA 2200002D01 gene |
2947 |
0.13 |
| chr1_183790757_183790934 | 0.33 |
Gm34068 |
predicted gene, 34068 |
41868 |
0.18 |
| chr2_166467245_166467396 | 0.33 |
5031425F14Rik |
RIKEN cDNA 5031425F14 gene |
19869 |
0.19 |
| chr1_71634492_71634670 | 0.33 |
Fn1 |
fibronectin 1 |
6707 |
0.2 |
| chr19_4436418_4436609 | 0.33 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
2490 |
0.17 |
| chr7_98572441_98572789 | 0.33 |
A630091E08Rik |
RIKEN cDNA A630091E08 gene |
9625 |
0.16 |
| chr10_61929391_61929572 | 0.32 |
Gm5750 |
predicted gene 5750 |
30143 |
0.16 |
| chr15_99792342_99792511 | 0.32 |
Lima1 |
LIM domain and actin binding 1 |
8453 |
0.08 |
| chr2_70421553_70421719 | 0.32 |
Rpl9-ps7 |
ribosomal protein L9, pseudogene 7 |
37346 |
0.14 |
| chr14_97847704_97847876 | 0.32 |
Gm25831 |
predicted gene, 25831 |
120201 |
0.06 |
| chr5_134629599_134629763 | 0.32 |
Eif4h |
eukaryotic translation initiation factor 4H |
9621 |
0.11 |
| chr5_32155495_32155671 | 0.32 |
Fosl2 |
fos-like antigen 2 |
9062 |
0.15 |
| chrX_43495267_43495434 | 0.32 |
Gm14588 |
predicted gene 14588 |
21396 |
0.21 |
| chr6_31100129_31100294 | 0.31 |
Gm13833 |
predicted gene 13833 |
2635 |
0.17 |
| chr13_59832569_59832762 | 0.31 |
Gm34961 |
predicted gene, 34961 |
9399 |
0.11 |
| chr11_96324315_96324468 | 0.31 |
Hoxb3 |
homeobox B3 |
1065 |
0.25 |
| chr3_37642128_37642470 | 0.31 |
Gm42922 |
predicted gene 42922 |
1643 |
0.21 |
| chr6_42221324_42221475 | 0.31 |
Tas2r144 |
taste receptor, type 2, member 144 |
6071 |
0.14 |
| chr19_54834637_54834803 | 0.31 |
Gm50191 |
predicted gene, 50191 |
102004 |
0.08 |
| chr15_82374486_82374674 | 0.31 |
Cyp2d22 |
cytochrome P450, family 2, subfamily d, polypeptide 22 |
742 |
0.37 |
| chr7_16287899_16288054 | 0.31 |
Ccdc9 |
coiled-coil domain containing 9 |
1181 |
0.32 |
| chr4_44994891_44995176 | 0.31 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
13533 |
0.1 |
| chr3_67551330_67551501 | 0.30 |
Gm35299 |
predicted gene, 35299 |
2010 |
0.23 |
| chr1_152032225_152032416 | 0.30 |
1700025G04Rik |
RIKEN cDNA 1700025G04 gene |
22972 |
0.23 |
| chr3_142707130_142707285 | 0.30 |
Kyat3 |
kynurenine aminotransferase 3 |
6113 |
0.15 |
| chr11_100855277_100855460 | 0.30 |
Stat5a |
signal transducer and activator of transcription 5A |
3983 |
0.16 |
| chr11_109559331_109559545 | 0.30 |
Arsg |
arylsulfatase G |
15684 |
0.15 |
| chr10_80136261_80136428 | 0.30 |
Cbarp |
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
756 |
0.4 |
| chr12_109512584_109512771 | 0.30 |
Gm34081 |
predicted gene, 34081 |
4063 |
0.07 |
| chr13_94063212_94063363 | 0.29 |
Lhfpl2 |
lipoma HMGIC fusion partner-like 2 |
5455 |
0.2 |
| chr14_16575472_16575639 | 0.29 |
Rarb |
retinoic acid receptor, beta |
12 |
0.52 |
| chr9_61164927_61165078 | 0.29 |
I730028E13Rik |
RIKEN cDNA I730028E13 gene |
26487 |
0.14 |
| chr1_174748737_174748905 | 0.29 |
Fmn2 |
formin 2 |
34495 |
0.22 |
| chr1_9645605_9645800 | 0.29 |
Gm29520 |
predicted gene 29520 |
5120 |
0.16 |
| chr4_128648691_128648859 | 0.29 |
Phc2 |
polyhomeotic 2 |
5927 |
0.17 |
| chrX_11630030_11630183 | 0.29 |
Gm14515 |
predicted gene 14515 |
28752 |
0.21 |
| chr2_104663420_104663722 | 0.29 |
Cstf3 |
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
95 |
0.96 |
| chr2_105235818_105235986 | 0.28 |
Them7 |
thioesterase superfamily member 7 |
11560 |
0.24 |
| chr11_51619305_51619469 | 0.28 |
Nhp2 |
NHP2 ribonucleoprotein |
348 |
0.8 |
| chr10_8071988_8072183 | 0.28 |
Gm48614 |
predicted gene, 48614 |
50793 |
0.15 |
| chr16_38286019_38286177 | 0.28 |
Nr1i2 |
nuclear receptor subfamily 1, group I, member 2 |
8726 |
0.15 |
| chr18_34927551_34927712 | 0.28 |
Etf1 |
eukaryotic translation termination factor 1 |
4376 |
0.14 |
| chr11_110032630_110032781 | 0.28 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
2624 |
0.32 |
| chr8_115968249_115968411 | 0.28 |
Gm45733 |
predicted gene 45733 |
3741 |
0.37 |
| chr12_83087732_83087912 | 0.27 |
Gm29530 |
predicted gene 29530 |
11052 |
0.17 |
| chr9_118655096_118655280 | 0.27 |
Itga9 |
integrin alpha 9 |
30647 |
0.17 |
| chr10_97387218_97387458 | 0.27 |
Mir3966 |
microRNA 3966 |
36140 |
0.19 |
| chr9_109090535_109090842 | 0.27 |
Plxnb1 |
plexin B1 |
4701 |
0.1 |
| chr16_18715486_18715659 | 0.27 |
Rps2-ps7 |
ribosomal protein S2, pseudogene 7 |
33356 |
0.12 |
| chr14_46419483_46419645 | 0.27 |
Gm15221 |
predicted gene 15221 |
17291 |
0.12 |
| chr17_8305079_8305235 | 0.27 |
4930506C21Rik |
RIKEN cDNA 4930506C21 gene |
5895 |
0.12 |
| chr4_118233934_118234146 | 0.27 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
1976 |
0.29 |
| chr12_25132357_25132520 | 0.27 |
Gm17746 |
predicted gene, 17746 |
3264 |
0.21 |
| chr3_148935992_148936150 | 0.27 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
18564 |
0.22 |
| chr11_16920982_16921140 | 0.27 |
Egfr |
epidermal growth factor receptor |
15876 |
0.16 |
| chr4_41524935_41525096 | 0.27 |
Fam219a |
family with sequence similarity 219, member A |
245 |
0.85 |
| chr3_122513095_122513259 | 0.27 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
955 |
0.47 |
| chr7_111020937_111021103 | 0.27 |
Ctr9 |
CTR9 homolog, Paf1/RNA polymerase II complex component |
7931 |
0.17 |
| chr14_26557730_26557942 | 0.27 |
Gm18215 |
predicted gene, 18215 |
5240 |
0.13 |
| chr8_104375235_104375402 | 0.27 |
Gm45877 |
predicted gene 45877 |
11650 |
0.09 |
| chr9_70827748_70828187 | 0.27 |
Gm3436 |
predicted pseudogene 3436 |
24609 |
0.16 |
| chr15_77072900_77073074 | 0.26 |
Gm24753 |
predicted gene, 24753 |
5196 |
0.12 |
| chr6_91111481_91112156 | 0.26 |
Nup210 |
nucleoporin 210 |
4978 |
0.17 |
| chr6_128549526_128549695 | 0.26 |
A2ml1 |
alpha-2-macroglobulin like 1 |
3668 |
0.12 |
| chr1_183414419_183414590 | 0.26 |
Gm37986 |
predicted gene, 37986 |
38 |
0.96 |
| chr12_110219862_110220027 | 0.26 |
Gm40576 |
predicted gene, 40576 |
16418 |
0.11 |
| chr7_110060209_110060360 | 0.26 |
Zfp143 |
zinc finger protein 143 |
933 |
0.32 |
| chr11_100318043_100318206 | 0.26 |
Eif1 |
eukaryotic translation initiation factor 1 |
1761 |
0.17 |
| chr10_28422676_28422859 | 0.26 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
137384 |
0.05 |
| chr10_120384864_120385074 | 0.26 |
9230105E05Rik |
RIKEN cDNA 9230105E05 gene |
4554 |
0.2 |
| chrX_163768401_163768583 | 0.26 |
Rnf138rt1 |
ring finger protein 138, retrogene 1 |
7163 |
0.23 |
| chr15_68255673_68255836 | 0.26 |
Zfat |
zinc finger and AT hook domain containing |
3033 |
0.26 |
| chr1_165679726_165679916 | 0.26 |
Gm44458 |
predicted gene, 44458 |
14944 |
0.1 |
| chr5_105239714_105239885 | 0.26 |
Gbp10 |
guanylate-binding protein 10 |
266 |
0.92 |
| chr4_35134107_35134262 | 0.26 |
Ifnk |
interferon kappa |
17872 |
0.17 |
| chr14_70207050_70207221 | 0.26 |
Sorbs3 |
sorbin and SH3 domain containing 3 |
502 |
0.48 |
| chr6_49150440_49150597 | 0.26 |
Gm18010 |
predicted gene, 18010 |
22241 |
0.13 |
| chr1_191449557_191449745 | 0.26 |
Gm32200 |
predicted gene, 32200 |
15397 |
0.13 |
| chr2_133495524_133495962 | 0.25 |
Gm14100 |
predicted gene 14100 |
32625 |
0.16 |
| chr11_94589492_94589674 | 0.25 |
Acsf2 |
acyl-CoA synthetase family member 2 |
12186 |
0.11 |
| chr6_144759587_144759981 | 0.25 |
Sox5 |
SRY (sex determining region Y)-box 5 |
22135 |
0.15 |
| chr10_99555138_99555310 | 0.25 |
Gm48085 |
predicted gene, 48085 |
17155 |
0.18 |
| chr7_16830530_16830865 | 0.25 |
Strn4 |
striatin, calmodulin binding protein 4 |
402 |
0.73 |
| chr13_68226599_68226750 | 0.25 |
AA414992 |
expressed sequence AA414992 |
45380 |
0.16 |
| chr7_45711383_45711838 | 0.25 |
Sphk2 |
sphingosine kinase 2 |
1833 |
0.12 |
| chr8_66579826_66579979 | 0.25 |
Gm16330 |
predicted gene 16330 |
9106 |
0.22 |
| chr10_92081958_92082460 | 0.25 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
6580 |
0.22 |
| chr11_86921503_86921665 | 0.25 |
Ypel2 |
yippee like 2 |
50440 |
0.12 |
| chr16_20674169_20674320 | 0.25 |
Eif4g1 |
eukaryotic translation initiation factor 4, gamma 1 |
133 |
0.89 |
| chr10_28167171_28167473 | 0.25 |
Gm22370 |
predicted gene, 22370 |
46799 |
0.17 |
| chr3_133534210_133534372 | 0.24 |
Tet2 |
tet methylcytosine dioxygenase 2 |
9995 |
0.17 |
| chr9_108913748_108913914 | 0.24 |
Tmem89 |
transmembrane protein 89 |
788 |
0.4 |
| chr14_69392700_69392874 | 0.24 |
Gm21451 |
predicted gene, 21451 |
1933 |
0.21 |
| chr18_5186924_5187075 | 0.24 |
Gm26682 |
predicted gene, 26682 |
21268 |
0.24 |
| chr11_75462092_75462507 | 0.24 |
Tlcd2 |
TLC domain containing 2 |
584 |
0.35 |
| chr1_55053215_55053405 | 0.24 |
E330011M16Rik |
RIKEN cDNA E330011M16 gene |
198 |
0.72 |
| chr1_36080918_36081086 | 0.24 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
12602 |
0.14 |
| chr16_22900205_22900497 | 0.24 |
Ahsg |
alpha-2-HS-glycoprotein |
5487 |
0.13 |
| chr8_22637028_22637191 | 0.24 |
Dkk4 |
dickkopf WNT signaling pathway inhibitor 4 |
13066 |
0.13 |
| chr19_32543108_32543270 | 0.24 |
Gm36419 |
predicted gene, 36419 |
159 |
0.96 |
| chr11_7196610_7196788 | 0.24 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1083 |
0.48 |
| chr8_69887766_69887933 | 0.24 |
Cilp2 |
cartilage intermediate layer protein 2 |
162 |
0.69 |
| chr11_16742202_16742481 | 0.24 |
Gm25698 |
predicted gene, 25698 |
9630 |
0.19 |
| chr4_139383343_139383534 | 0.24 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
2769 |
0.16 |
| chr9_74931997_74932158 | 0.23 |
Fam214a |
family with sequence similarity 214, member A |
20807 |
0.17 |
| chr2_128179604_128179779 | 0.23 |
Gm14009 |
predicted gene 14009 |
10296 |
0.22 |
| chr5_122882739_122882908 | 0.23 |
Kdm2b |
lysine (K)-specific demethylase 2B |
590 |
0.67 |
| chr2_5600300_5600460 | 0.23 |
Gm13216 |
predicted gene 13216 |
3191 |
0.34 |
| chr19_60828743_60828894 | 0.23 |
Fam45a |
family with sequence similarity 45, member A |
1734 |
0.25 |
| chr9_25570473_25570647 | 0.23 |
Gm25346 |
predicted gene, 25346 |
755 |
0.7 |
| chr12_105784205_105784562 | 0.23 |
Papola |
poly (A) polymerase alpha |
311 |
0.9 |
| chr3_38589085_38589252 | 0.23 |
Gm43538 |
predicted gene 43538 |
25079 |
0.16 |
| chr11_120543038_120543310 | 0.23 |
Mcrip1 |
MAPK regulated corepressor interacting protein 1 |
6527 |
0.06 |
| chr8_70552951_70553110 | 0.23 |
Ell |
elongation factor RNA polymerase II |
13342 |
0.08 |
| chr13_74769953_74770127 | 0.23 |
Cast |
calpastatin |
550 |
0.67 |
| chr2_148443567_148443727 | 0.23 |
Cd93 |
CD93 antigen |
84 |
0.97 |
| chr6_31323903_31324093 | 0.23 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
2027 |
0.28 |
| chr11_106967758_106967936 | 0.23 |
Gm11707 |
predicted gene 11707 |
5243 |
0.15 |
| chr6_115480476_115480638 | 0.23 |
Gm44079 |
predicted gene, 44079 |
15567 |
0.16 |
| chr4_140770556_140770730 | 0.23 |
Padi4 |
peptidyl arginine deiminase, type IV |
3593 |
0.16 |
| chr4_106564660_106564819 | 0.23 |
Dhcr24 |
24-dehydrocholesterol reductase |
3701 |
0.14 |
| chr15_80720254_80720552 | 0.23 |
Gm49512 |
predicted gene, 49512 |
8132 |
0.15 |
| chr4_60424050_60424352 | 0.23 |
Mup9 |
major urinary protein 9 |
2249 |
0.23 |
| chr4_148085280_148085458 | 0.23 |
Agtrap |
angiotensin II, type I receptor-associated protein |
593 |
0.56 |
| chr13_93177685_93177846 | 0.23 |
Tent2 |
terminal nucleotidyltransferase 2 |
9336 |
0.2 |
| chr1_10099708_10099862 | 0.22 |
Cspp1 |
centrosome and spindle pole associated protein 1 |
3921 |
0.2 |
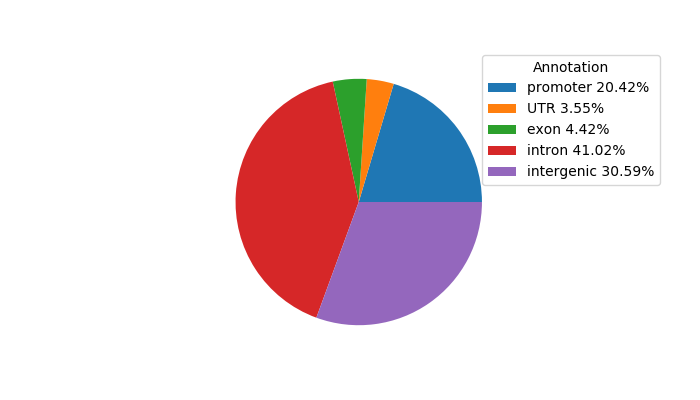
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.1 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.2 | GO:0006524 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.2 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.0 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.0 | GO:0003133 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.0 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.0 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) |
| 0.0 | 0.1 | GO:0006056 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.0 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.0 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.4 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0018568 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.1 | GO:0018586 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |