Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
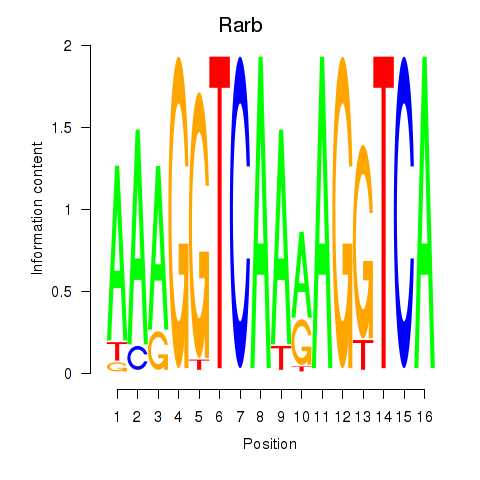
Results for Rarb
Z-value: 1.93
Transcription factors associated with Rarb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarb
|
ENSMUSG00000017491.8 | retinoic acid receptor, beta |
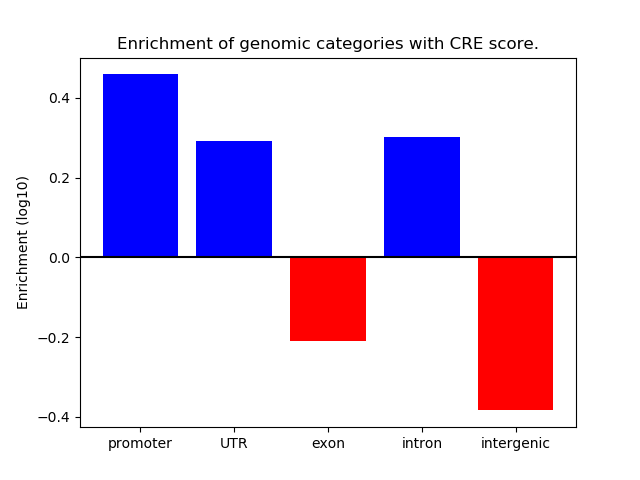
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_16573210_16573389 | Rarb | 1746 | 0.364795 | -0.96 | 2.7e-03 | Click! |
| chr14_16572962_16573173 | Rarb | 1978 | 0.340832 | -0.95 | 3.0e-03 | Click! |
| chr14_16570330_16570481 | Rarb | 4640 | 0.249769 | -0.94 | 5.4e-03 | Click! |
| chr14_16735456_16735607 | Rarb | 83474 | 0.100150 | 0.93 | 7.4e-03 | Click! |
| chr14_16575472_16575639 | Rarb | 12 | 0.517456 | 0.90 | 1.4e-02 | Click! |
Activity of the Rarb motif across conditions
Conditions sorted by the z-value of the Rarb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
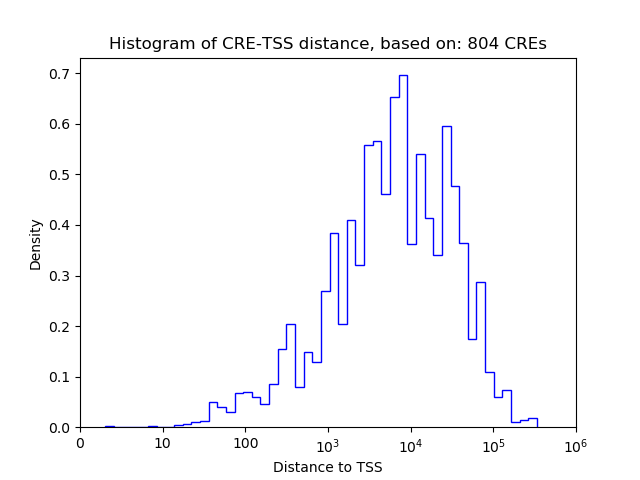
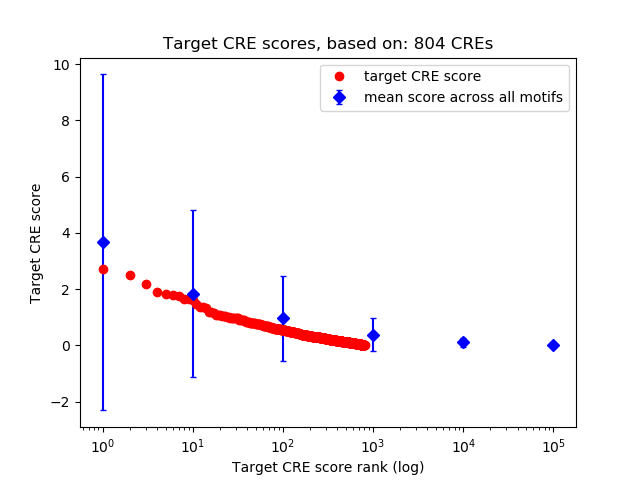
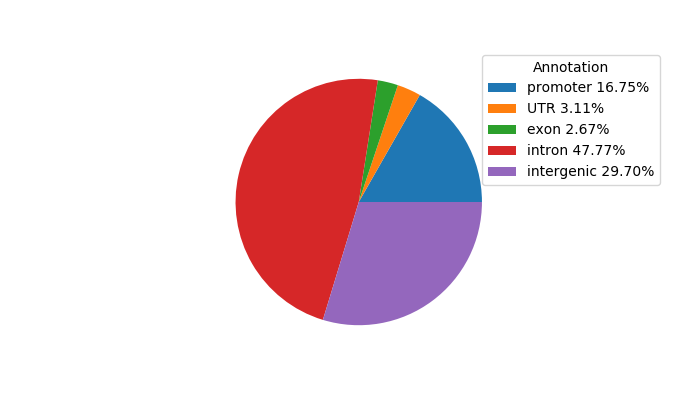
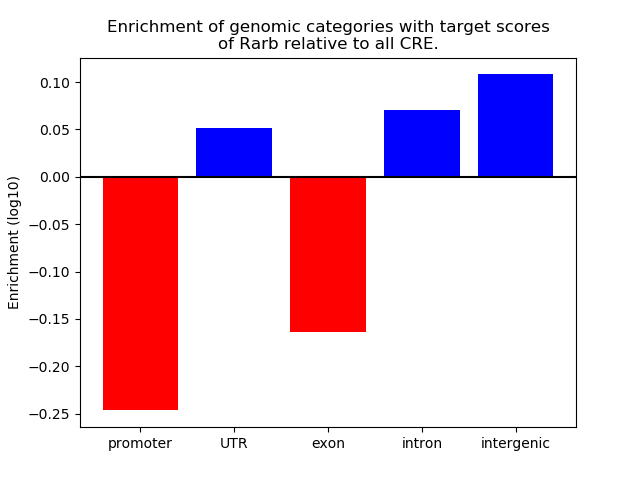
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_71429918_71430089 | 2.70 |
1700083H02Rik |
RIKEN cDNA 1700083H02 gene |
45795 |
0.12 |
| chr8_94217878_94218059 | 2.51 |
Nup93 |
nucleoporin 93 |
3362 |
0.14 |
| chr4_150287636_150287787 | 2.18 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
5899 |
0.16 |
| chr6_89138858_89139017 | 1.91 |
Gm1965 |
predicted gene 1965 |
2057 |
0.29 |
| chr11_50733265_50733435 | 1.83 |
Gm12200 |
predicted gene 12200 |
7806 |
0.16 |
| chr2_114776001_114776450 | 1.78 |
Gm13975 |
predicted gene 13975 |
68113 |
0.11 |
| chr3_104541230_104541625 | 1.77 |
Lrig2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
29509 |
0.1 |
| chr4_43234117_43234284 | 1.66 |
Unc13b |
unc-13 homolog B |
393 |
0.84 |
| chr5_118292276_118292427 | 1.66 |
Gm25076 |
predicted gene, 25076 |
25902 |
0.16 |
| chr14_51102456_51102668 | 1.60 |
Rnase4 |
ribonuclease, RNase A family 4 |
6203 |
0.08 |
| chr8_115644414_115644565 | 1.48 |
Gm23677 |
predicted gene, 23677 |
53216 |
0.13 |
| chr5_88969041_88969210 | 1.38 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
34553 |
0.2 |
| chr19_21038157_21038331 | 1.36 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
66258 |
0.11 |
| chr8_124345179_124345346 | 1.32 |
Gm24459 |
predicted gene, 24459 |
11976 |
0.14 |
| chr5_146107597_146107748 | 1.19 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
28405 |
0.11 |
| chr19_47385384_47385539 | 1.17 |
Sh3pxd2a |
SH3 and PX domains 2A |
24898 |
0.18 |
| chr17_56470625_56470795 | 1.16 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
3917 |
0.18 |
| chr5_121730204_121730378 | 1.08 |
Gm16552 |
predicted gene 16552 |
2888 |
0.16 |
| chr13_40913304_40913608 | 1.06 |
Gm48188 |
predicted gene, 48188 |
1249 |
0.3 |
| chr10_84055512_84055676 | 1.06 |
Gm37908 |
predicted gene, 37908 |
6164 |
0.21 |
| chr13_79286283_79286457 | 1.05 |
Gm29318 |
predicted gene 29318 |
658 |
0.76 |
| chr4_3736363_3736519 | 1.04 |
Gm11805 |
predicted gene 11805 |
15415 |
0.15 |
| chr9_109115339_109115519 | 1.03 |
Plxnb1 |
plexin B1 |
8635 |
0.1 |
| chrX_42066716_42066867 | 1.02 |
Xiap |
X-linked inhibitor of apoptosis |
1045 |
0.59 |
| chrX_101391513_101391680 | 0.99 |
Gjb1 |
gap junction protein, beta 1 |
7860 |
0.14 |
| chr2_69415195_69415364 | 0.99 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
34834 |
0.16 |
| chr1_75662127_75662446 | 0.99 |
Gm5257 |
predicted gene 5257 |
25896 |
0.15 |
| chr5_64883553_64883984 | 0.99 |
AC161757.1 |
toll-like receptor 1 (Tlr1) pseudogene |
15201 |
0.15 |
| chr8_39639786_39639944 | 0.98 |
Msr1 |
macrophage scavenger receptor 1 |
2755 |
0.39 |
| chr5_122693537_122693811 | 0.98 |
Gm10064 |
predicted gene 10064 |
3926 |
0.14 |
| chr4_141280593_141280762 | 0.97 |
Gm13056 |
predicted gene 13056 |
2238 |
0.18 |
| chr17_24797110_24797261 | 0.96 |
Meiob |
meiosis specific with OB domains |
7104 |
0.09 |
| chr10_34136560_34136736 | 0.91 |
Calhm6 |
calcium homeostasis modulator family member 6 |
8664 |
0.13 |
| chr15_26377246_26377425 | 0.91 |
Marchf11 |
membrane associated ring-CH-type finger 11 |
10384 |
0.3 |
| chr17_26687116_26687267 | 0.90 |
Atp6v0e |
ATPase, H+ transporting, lysosomal V0 subunit E |
10779 |
0.14 |
| chr5_145982849_145983558 | 0.90 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
8440 |
0.13 |
| chr5_137481993_137482153 | 0.89 |
Epo |
erythropoietin |
3743 |
0.1 |
| chr7_16229973_16230150 | 0.86 |
Gm45409 |
predicted gene 45409 |
1161 |
0.3 |
| chr7_80464871_80465056 | 0.84 |
Blm |
Bloom syndrome, RecQ like helicase |
1086 |
0.43 |
| chr14_28283569_28283764 | 0.84 |
Gm20242 |
predicted gene, 20242 |
132331 |
0.05 |
| chr6_124666813_124667013 | 0.83 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
3698 |
0.1 |
| chr18_46335696_46335862 | 0.83 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
4650 |
0.16 |
| chr4_44051303_44051454 | 0.80 |
Gne |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
3919 |
0.17 |
| chr10_79887085_79887245 | 0.80 |
Elane |
elastase, neutrophil expressed |
918 |
0.23 |
| chr12_72763283_72763641 | 0.79 |
Ppm1a |
protein phosphatase 1A, magnesium dependent, alpha isoform |
2147 |
0.31 |
| chr3_95694014_95694171 | 0.79 |
Adamtsl4 |
ADAMTS-like 4 |
6175 |
0.11 |
| chr10_126691394_126691560 | 0.78 |
Mir378d |
microRNA 378d |
18914 |
0.15 |
| chr12_71875530_71876050 | 0.78 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
13940 |
0.21 |
| chr10_94810057_94810242 | 0.78 |
Gm24186 |
predicted gene, 24186 |
11506 |
0.16 |
| chr1_191821111_191821273 | 0.77 |
Nek2 |
NIMA (never in mitosis gene a)-related expressed kinase 2 |
252 |
0.8 |
| chr13_55103446_55103605 | 0.77 |
Zfp346 |
zinc finger protein 346 |
1786 |
0.23 |
| chr13_93622998_93623302 | 0.76 |
Gm15622 |
predicted gene 15622 |
2232 |
0.26 |
| chr15_98915891_98916274 | 0.76 |
Lmbr1l |
limb region 1 like |
130 |
0.89 |
| chr4_32160991_32161147 | 0.76 |
Gm11928 |
predicted gene 11928 |
11283 |
0.21 |
| chr5_16317582_16317767 | 0.75 |
Cacna2d1 |
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
8311 |
0.25 |
| chr11_31933098_31933283 | 0.74 |
4930524B15Rik |
RIKEN cDNA 4930524B15 gene |
32402 |
0.16 |
| chr2_29483450_29483627 | 0.73 |
Mir133c |
microRNA 133c |
8244 |
0.15 |
| chr12_35739125_35739291 | 0.73 |
Gm48264 |
predicted gene, 48264 |
7021 |
0.18 |
| chr3_104214062_104214367 | 0.73 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
5889 |
0.12 |
| chr14_55899299_55899513 | 0.72 |
Sdr39u1 |
short chain dehydrogenase/reductase family 39U, member 1 |
449 |
0.66 |
| chr8_117337291_117337451 | 0.70 |
Cmip |
c-Maf inducing protein |
11799 |
0.25 |
| chr6_85864689_85864938 | 0.70 |
Nat8f2 |
N-acetyltransferase 8 (GCN5-related) family member 2 |
4345 |
0.1 |
| chr8_11148911_11149083 | 0.70 |
Gm44717 |
predicted gene 44717 |
249 |
0.91 |
| chr17_29487122_29487273 | 0.68 |
Pim1 |
proviral integration site 1 |
3556 |
0.13 |
| chr5_43156906_43157079 | 0.68 |
Gm42552 |
predicted gene 42552 |
8665 |
0.21 |
| chr1_131970433_131970601 | 0.68 |
Slc45a3 |
solute carrier family 45, member 3 |
92 |
0.95 |
| chr17_73072435_73072612 | 0.67 |
Lclat1 |
lysocardiolipin acyltransferase 1 |
35462 |
0.16 |
| chr4_141287858_141288020 | 0.67 |
Gm13056 |
predicted gene 13056 |
9500 |
0.1 |
| chr14_36971294_36971453 | 0.67 |
Ccser2 |
coiled-coil serine rich 2 |
2596 |
0.29 |
| chr3_131489084_131489522 | 0.66 |
Sgms2 |
sphingomyelin synthase 2 |
1176 |
0.58 |
| chr5_88953989_88954155 | 0.65 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
19500 |
0.24 |
| chr11_108254783_108254943 | 0.64 |
Gm11655 |
predicted gene 11655 |
73013 |
0.09 |
| chr2_84762961_84763125 | 0.64 |
Serping1 |
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
12085 |
0.08 |
| chr15_47749931_47750085 | 0.63 |
Gm16300 |
predicted gene 16300 |
46181 |
0.17 |
| chr7_68280697_68280888 | 0.61 |
Gm16157 |
predicted gene 16157 |
4198 |
0.16 |
| chr11_83409286_83409449 | 0.61 |
Rasl10b |
RAS-like, family 10, member B |
230 |
0.88 |
| chr9_15442312_15442468 | 0.61 |
Gm48634 |
predicted gene, 48634 |
1919 |
0.24 |
| chr18_76051312_76051463 | 0.60 |
Zbtb7c |
zinc finger and BTB domain containing 7C |
8071 |
0.22 |
| chr10_77109671_77109843 | 0.60 |
Col18a1 |
collagen, type XVIII, alpha 1 |
3948 |
0.21 |
| chr15_8968437_8968609 | 0.60 |
Ranbp3l |
RAN binding protein 3-like |
97 |
0.97 |
| chr7_112121062_112121238 | 0.59 |
Usp47 |
ubiquitin specific peptidase 47 |
30444 |
0.19 |
| chr11_86989298_86989461 | 0.59 |
Ypel2 |
yippee like 2 |
4328 |
0.19 |
| chr6_72212736_72212914 | 0.59 |
Atoh8 |
atonal bHLH transcription factor 8 |
21712 |
0.14 |
| chr17_24478264_24478594 | 0.59 |
Mlst8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
49 |
0.92 |
| chr8_69063559_69063737 | 0.59 |
Slc18a1 |
solute carrier family 18 (vesicular monoamine), member 1 |
11860 |
0.15 |
| chr2_38925242_38925421 | 0.59 |
Nr6a1 |
nuclear receptor subfamily 6, group A, member 1 |
886 |
0.43 |
| chr11_69842335_69842486 | 0.58 |
Tmem256 |
transmembrane protein 256 |
3772 |
0.07 |
| chr4_133338574_133338734 | 0.58 |
Wdtc1 |
WD and tetratricopeptide repeats 1 |
617 |
0.65 |
| chr9_70248725_70249019 | 0.58 |
Myo1e |
myosin IE |
41504 |
0.15 |
| chr8_33904851_33905143 | 0.57 |
Rbpms |
RNA binding protein gene with multiple splicing |
13233 |
0.17 |
| chr7_101465911_101466071 | 0.57 |
Pde2a |
phosphodiesterase 2A, cGMP-stimulated |
5047 |
0.16 |
| chr16_24154494_24154889 | 0.57 |
Gm32126 |
predicted gene, 32126 |
13870 |
0.2 |
| chr2_25266466_25266742 | 0.57 |
Tprn |
taperin |
960 |
0.23 |
| chr2_103811926_103812077 | 0.56 |
Gm13878 |
predicted gene 13878 |
384 |
0.68 |
| chr10_63226846_63227134 | 0.56 |
Herc4 |
hect domain and RLD 4 |
16820 |
0.11 |
| chr11_95038769_95038930 | 0.56 |
Pdk2 |
pyruvate dehydrogenase kinase, isoenzyme 2 |
1709 |
0.23 |
| chr9_83866290_83866453 | 0.55 |
Ttk |
Ttk protein kinase |
31605 |
0.14 |
| chr6_129175697_129175889 | 0.55 |
Gm26160 |
predicted gene, 26160 |
973 |
0.41 |
| chr2_113537620_113538205 | 0.55 |
Gm13964 |
predicted gene 13964 |
33878 |
0.16 |
| chr4_130112167_130112332 | 0.55 |
Pef1 |
penta-EF hand domain containing 1 |
4693 |
0.16 |
| chr11_106502645_106502807 | 0.54 |
Gm22711 |
predicted gene, 22711 |
1481 |
0.29 |
| chr5_134279549_134279704 | 0.54 |
Gtf2i |
general transcription factor II I |
239 |
0.89 |
| chr6_95819185_95819380 | 0.54 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
100482 |
0.08 |
| chr9_35136038_35136227 | 0.54 |
Gm24262 |
predicted gene, 24262 |
8504 |
0.12 |
| chr18_67958643_67958805 | 0.54 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
25467 |
0.19 |
| chrX_155337365_155337828 | 0.53 |
Prdx4 |
peroxiredoxin 4 |
871 |
0.59 |
| chr5_121665076_121665253 | 0.52 |
Brap |
BRCA1 associated protein |
1362 |
0.3 |
| chr14_20638160_20638341 | 0.52 |
2810402E24Rik |
RIKEN cDNA 2810402E24 gene |
2759 |
0.13 |
| chr4_119259446_119259606 | 0.51 |
Gm12927 |
predicted gene 12927 |
2551 |
0.13 |
| chr8_121562702_121562861 | 0.51 |
Fbxo31 |
F-box protein 31 |
2818 |
0.16 |
| chr5_113735107_113735259 | 0.51 |
Ficd |
FIC domain containing |
620 |
0.6 |
| chr11_120726590_120726778 | 0.51 |
Dcxr |
dicarbonyl L-xylulose reductase |
545 |
0.48 |
| chr14_122765965_122766143 | 0.51 |
Pcca |
propionyl-Coenzyme A carboxylase, alpha polypeptide |
24345 |
0.21 |
| chr5_134170407_134170687 | 0.50 |
Rcc1l |
reculator of chromosome condensation 1 like |
1172 |
0.37 |
| chr4_140636700_140636879 | 0.50 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
11961 |
0.16 |
| chr2_121968638_121968832 | 0.49 |
Ctdspl2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
347 |
0.85 |
| chr15_36904059_36904218 | 0.49 |
Gm10384 |
predicted gene 10384 |
24322 |
0.13 |
| chr3_108804290_108804468 | 0.49 |
Stxbp3 |
syntaxin binding protein 3 |
1352 |
0.39 |
| chr9_116947417_116947569 | 0.49 |
Gm18489 |
predicted gene, 18489 |
111665 |
0.07 |
| chr17_20287010_20287161 | 0.49 |
Vmn2r106 |
vomeronasal 2, receptor 106 |
1532 |
0.35 |
| chr10_121455166_121455317 | 0.48 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
6972 |
0.13 |
| chr15_64273225_64273401 | 0.47 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
38376 |
0.15 |
| chr2_31530318_31530475 | 0.47 |
Ass1 |
argininosuccinate synthetase 1 |
11906 |
0.15 |
| chr15_36914757_36914955 | 0.47 |
Gm41300 |
predicted gene, 41300 |
16830 |
0.15 |
| chr7_141337790_141337972 | 0.47 |
Eps8l2 |
EPS8-like 2 |
999 |
0.3 |
| chr16_4757027_4757557 | 0.46 |
Hmox2 |
heme oxygenase 2 |
298 |
0.84 |
| chr5_74813446_74813635 | 0.46 |
Gm42575 |
predicted gene 42575 |
63658 |
0.09 |
| chr13_28757565_28757740 | 0.46 |
Mir6368 |
microRNA 6368 |
46779 |
0.15 |
| chr11_21058339_21058501 | 0.46 |
Peli1 |
pellino 1 |
32871 |
0.16 |
| chr11_70745785_70745982 | 0.46 |
Gm12320 |
predicted gene 12320 |
9268 |
0.08 |
| chr15_102171688_102171839 | 0.46 |
Csad |
cysteine sulfinic acid decarboxylase |
7256 |
0.1 |
| chr7_123850432_123850600 | 0.46 |
Gm23788 |
predicted gene, 23788 |
91693 |
0.08 |
| chr8_71307755_71307928 | 0.46 |
Myo9b |
myosin IXb |
25976 |
0.11 |
| chr18_20938375_20938560 | 0.46 |
Rnf125 |
ring finger protein 125 |
6158 |
0.22 |
| chr11_75661487_75661650 | 0.45 |
Myo1c |
myosin IC |
165 |
0.92 |
| chr15_99790865_99791044 | 0.45 |
Lima1 |
LIM domain and actin binding 1 |
6981 |
0.09 |
| chr11_105237702_105237861 | 0.45 |
Tlk2 |
tousled-like kinase 2 (Arabidopsis) |
8977 |
0.19 |
| chrX_60831870_60832021 | 0.45 |
Gm14660 |
predicted gene 14660 |
13737 |
0.19 |
| chr5_81908094_81908272 | 0.45 |
2900064F13Rik |
RIKEN cDNA 2900064F13 gene |
81367 |
0.1 |
| chr7_80830498_80830680 | 0.44 |
Iqgap1 |
IQ motif containing GTPase activating protein 1 |
4615 |
0.18 |
| chr13_32169128_32169451 | 0.44 |
Gmds |
GDP-mannose 4, 6-dehydratase |
57952 |
0.13 |
| chr2_12060466_12060629 | 0.44 |
Gm13310 |
predicted gene 13310 |
23445 |
0.21 |
| chr17_47888408_47888575 | 0.43 |
Foxp4 |
forkhead box P4 |
58 |
0.96 |
| chr11_78698801_78698963 | 0.43 |
Nlk |
nemo like kinase |
1509 |
0.33 |
| chr11_44173581_44173732 | 0.43 |
Gm12154 |
predicted gene 12154 |
93553 |
0.08 |
| chr13_31171886_31172048 | 0.43 |
Gm11372 |
predicted gene 11372 |
27119 |
0.19 |
| chr7_134497184_134497335 | 0.43 |
D7Ertd443e |
DNA segment, Chr 7, ERATO Doi 443, expressed |
1725 |
0.51 |
| chr3_115682777_115683090 | 0.43 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
32139 |
0.17 |
| chr2_80600719_80600876 | 0.43 |
Gm13688 |
predicted gene 13688 |
1936 |
0.21 |
| chr4_11147830_11148134 | 0.42 |
Gm11830 |
predicted gene 11830 |
3272 |
0.15 |
| chr18_38205562_38205736 | 0.42 |
Pcdh1 |
protocadherin 1 |
1809 |
0.24 |
| chr16_21865351_21865519 | 0.41 |
Gm26744 |
predicted gene, 26744 |
19659 |
0.11 |
| chr7_110920925_110921090 | 0.41 |
Mrvi1 |
MRV integration site 1 |
2696 |
0.25 |
| chr5_149198922_149199082 | 0.41 |
Uspl1 |
ubiquitin specific peptidase like 1 |
7666 |
0.09 |
| chr2_151925877_151926031 | 0.40 |
Angpt4 |
angiopoietin 4 |
14744 |
0.13 |
| chr2_3770483_3770646 | 0.39 |
Fam107b |
family with sequence similarity 107, member B |
144 |
0.96 |
| chr17_88509210_88509380 | 0.39 |
Ppp1r21 |
protein phosphatase 1, regulatory subunit 21 |
20823 |
0.16 |
| chr18_60769849_60770150 | 0.39 |
Rps14 |
ribosomal protein S14 |
4597 |
0.15 |
| chr11_119954339_119954501 | 0.39 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
5601 |
0.13 |
| chr9_77927047_77927209 | 0.38 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
5196 |
0.17 |
| chr11_35783421_35783583 | 0.38 |
Mir103-1 |
microRNA 103-1 |
1106 |
0.43 |
| chr1_7104338_7104546 | 0.38 |
Pcmtd1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
15276 |
0.15 |
| chr16_23218528_23218719 | 0.38 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
6117 |
0.11 |
| chr6_49210657_49210827 | 0.38 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
3419 |
0.2 |
| chr10_108449802_108449991 | 0.38 |
Gm36283 |
predicted gene, 36283 |
3294 |
0.25 |
| chr9_45833631_45833782 | 0.38 |
Gm7286 |
predicted gene 7286 |
3229 |
0.12 |
| chr12_80146592_80146753 | 0.38 |
2310015A10Rik |
RIKEN cDNA 2310015A10 gene |
13828 |
0.12 |
| chr9_44406569_44406995 | 0.37 |
Trappc4 |
trafficking protein particle complex 4 |
260 |
0.51 |
| chr3_85786657_85786819 | 0.37 |
Fam160a1 |
family with sequence similarity 160, member A1 |
30553 |
0.16 |
| chr5_112344431_112344696 | 0.37 |
Hps4 |
HPS4, biogenesis of lysosomal organelles complex 3 subunit 2 |
1463 |
0.2 |
| chr2_75999812_75999973 | 0.37 |
Pde11a |
phosphodiesterase 11A |
6151 |
0.18 |
| chr10_14035731_14035911 | 0.37 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
68797 |
0.09 |
| chr5_107272625_107272804 | 0.37 |
Gm42900 |
predicted gene 42900 |
16210 |
0.12 |
| chr17_84026540_84026710 | 0.36 |
Gm35551 |
predicted gene, 35551 |
606 |
0.62 |
| chr7_5021714_5021881 | 0.36 |
Zfp865 |
zinc finger protein 865 |
1210 |
0.17 |
| chr18_24652431_24652590 | 0.36 |
Mocos |
molybdenum cofactor sulfurase |
1181 |
0.43 |
| chr12_100191588_100191746 | 0.35 |
Gm10433 |
predicted gene 10433 |
2894 |
0.18 |
| chr12_21136203_21136399 | 0.35 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
24347 |
0.18 |
| chr3_68593185_68593343 | 0.35 |
Schip1 |
schwannomin interacting protein 1 |
2911 |
0.3 |
| chr12_87408323_87408513 | 0.35 |
Gm8557 |
predicted gene 8557 |
12093 |
0.07 |
| chr14_63246048_63246212 | 0.35 |
Gata4 |
GATA binding protein 4 |
859 |
0.59 |
| chr8_111775967_111776118 | 0.35 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
32233 |
0.14 |
| chr1_36571715_36571905 | 0.35 |
D430040D24Rik |
RIKEN cDNA D430040D24 gene |
13451 |
0.09 |
| chrX_99138009_99138168 | 0.35 |
Efnb1 |
ephrin B1 |
43 |
0.98 |
| chr16_23592594_23592752 | 0.35 |
Rtp4 |
receptor transporter protein 4 |
9069 |
0.21 |
| chr6_124528028_124528336 | 0.35 |
Gm16567 |
predicted gene 16567 |
2729 |
0.14 |
| chr4_137471059_137471217 | 0.35 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
2335 |
0.21 |
| chr9_63534222_63534551 | 0.34 |
Gm16759 |
predicted gene, 16759 |
3034 |
0.28 |
| chr4_134488308_134488479 | 0.34 |
Gm13250 |
predicted gene 13250 |
4829 |
0.11 |
| chr6_116415099_116415265 | 0.34 |
Alox5 |
arachidonate 5-lipoxygenase |
1162 |
0.37 |
| chr5_111336024_111336199 | 0.34 |
Pitpnb |
phosphatidylinositol transfer protein, beta |
5304 |
0.19 |
| chr15_36585261_36585431 | 0.34 |
Gm44310 |
predicted gene, 44310 |
1802 |
0.26 |
| chr12_79689447_79689598 | 0.34 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
235211 |
0.02 |
| chr5_147880674_147880894 | 0.33 |
Slc46a3 |
solute carrier family 46, member 3 |
915 |
0.53 |
| chr10_18074695_18074946 | 0.33 |
Reps1 |
RalBP1 associated Eps domain containing protein |
16648 |
0.19 |
| chr7_98141341_98141502 | 0.33 |
Omp |
olfactory marker protein |
4081 |
0.18 |
| chr7_127811270_127811556 | 0.33 |
Stx1b |
syntaxin 1B |
489 |
0.54 |
| chr1_157526751_157526916 | 0.33 |
Sec16b |
SEC16 homolog B (S. cerevisiae) |
686 |
0.65 |
| chr5_107834436_107834622 | 0.33 |
Evi5 |
ecotropic viral integration site 5 |
2951 |
0.15 |
| chr13_98944666_98945270 | 0.32 |
Gm35215 |
predicted gene, 35215 |
948 |
0.45 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.2 | 0.6 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.6 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.3 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 1.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.2 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.3 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.5 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.2 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.0 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.0 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.4 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.2 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.1 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.3 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0052830 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0052760 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0044105 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.3 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.0 | GO:0019198 | transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.0 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |