Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
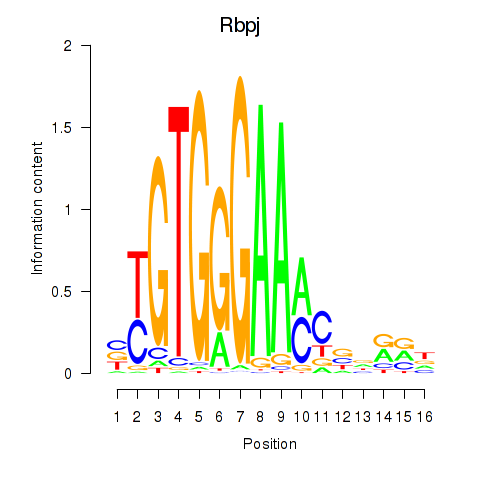
Results for Rbpj
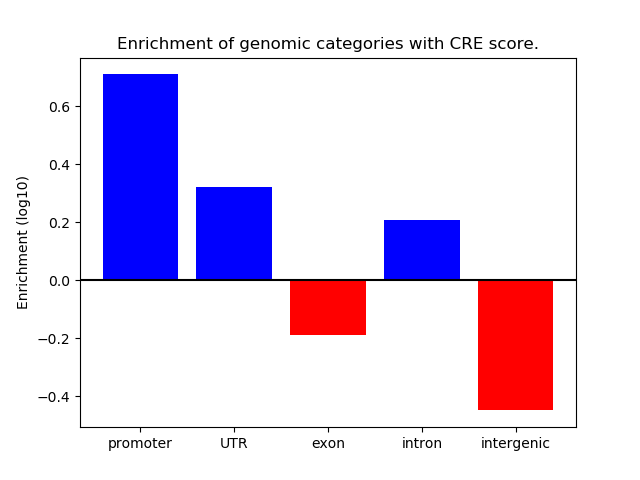
Z-value: 1.61
Transcription factors associated with Rbpj
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rbpj
|
ENSMUSG00000039191.6 | recombination signal binding protein for immunoglobulin kappa J region |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_53656339_53656508 | Rbpj | 10685 | 0.236867 | 0.85 | 3.4e-02 | Click! |
| chr5_53590620_53590774 | Rbpj | 226 | 0.946187 | 0.83 | 4.2e-02 | Click! |
| chr5_53469500_53469671 | Rbpj | 3433 | 0.255326 | 0.75 | 8.8e-02 | Click! |
| chr5_53589239_53589555 | Rbpj | 817 | 0.678983 | 0.71 | 1.1e-01 | Click! |
| chr5_53444100_53444263 | Rbpj | 21971 | 0.165880 | 0.70 | 1.2e-01 | Click! |
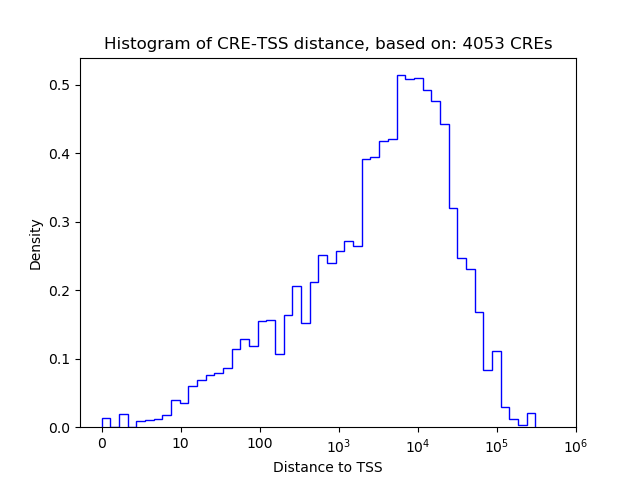
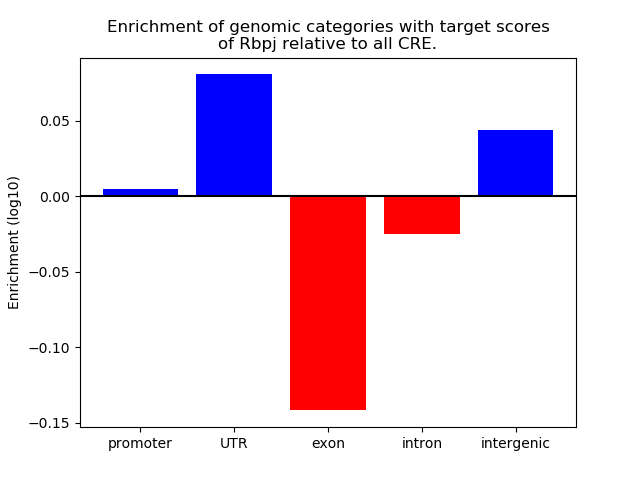
Activity of the Rbpj motif across conditions
Conditions sorted by the z-value of the Rbpj motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
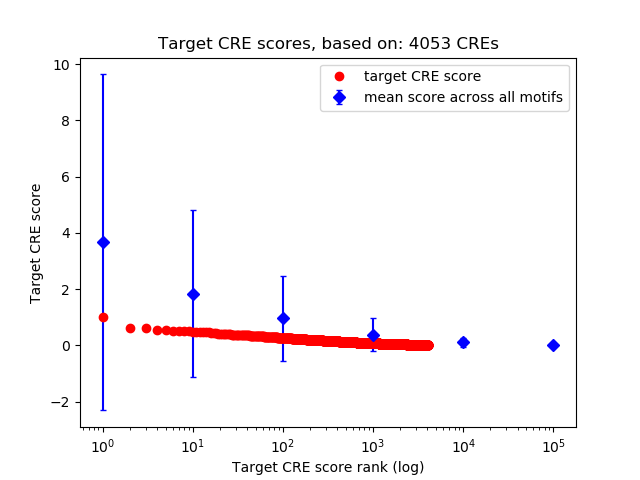
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_20989972_20990130 | 1.02 |
Rnf138 |
ring finger protein 138 |
11290 |
0.2 |
| chr19_23044990_23045152 | 0.62 |
Gm50136 |
predicted gene, 50136 |
16383 |
0.2 |
| chr13_111937084_111937265 | 0.62 |
Gm9025 |
predicted gene 9025 |
52997 |
0.09 |
| chr8_123657838_123658224 | 0.55 |
Rhou |
ras homolog family member U |
4102 |
0.05 |
| chr10_61337817_61337968 | 0.53 |
Pald1 |
phosphatase domain containing, paladin 1 |
4435 |
0.15 |
| chr8_117700284_117700470 | 0.50 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
1527 |
0.3 |
| chr6_144492068_144492244 | 0.50 |
Sox5os2 |
SRY (sex determining region Y)-box 5, opposite strand 2 |
10931 |
0.29 |
| chr4_123427592_123427944 | 0.49 |
Gm12924 |
predicted gene 12924 |
7849 |
0.18 |
| chr6_28929316_28929468 | 0.49 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
49349 |
0.12 |
| chr1_21305444_21305801 | 0.49 |
Gm4956 |
predicted gene 4956 |
7310 |
0.1 |
| chr17_27556354_27556505 | 0.48 |
Hmga1 |
high mobility group AT-hook 1 |
68 |
0.51 |
| chr11_58006609_58006789 | 0.47 |
Larp1 |
La ribonucleoprotein domain family, member 1 |
2365 |
0.25 |
| chr7_19326476_19326637 | 0.47 |
Gm44698 |
predicted gene 44698 |
6573 |
0.07 |
| chr1_136937150_136937320 | 0.47 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
3348 |
0.29 |
| chr8_36491389_36491571 | 0.46 |
Trmt9b |
tRNA methyltransferase 9B |
2289 |
0.34 |
| chr2_91264224_91264426 | 0.44 |
Arfgap2 |
ADP-ribosylation factor GTPase activating protein 2 |
649 |
0.5 |
| chr1_190076180_190076366 | 0.44 |
Gm28172 |
predicted gene 28172 |
92397 |
0.07 |
| chr11_103021982_103022133 | 0.42 |
Dcakd |
dephospho-CoA kinase domain containing |
4889 |
0.13 |
| chr3_98295816_98295967 | 0.42 |
Gm43189 |
predicted gene 43189 |
8903 |
0.15 |
| chr7_6696462_6696626 | 0.41 |
Zim1 |
zinc finger, imprinted 1 |
94 |
0.95 |
| chr12_102453656_102453840 | 0.41 |
Gm30198 |
predicted gene, 30198 |
6420 |
0.17 |
| chr13_92609818_92609992 | 0.41 |
Serinc5 |
serine incorporator 5 |
1186 |
0.52 |
| chr2_128731575_128731758 | 0.40 |
Gm14011 |
predicted gene 14011 |
22039 |
0.13 |
| chr10_11615264_11615424 | 0.39 |
Gm9797 |
predicted pseudogene 9797 |
6046 |
0.22 |
| chr15_100666435_100666616 | 0.39 |
Bin2 |
bridging integrator 2 |
2980 |
0.13 |
| chr6_142708472_142708687 | 0.39 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
6264 |
0.22 |
| chr11_70585533_70585714 | 0.38 |
Mink1 |
misshapen-like kinase 1 (zebrafish) |
13365 |
0.07 |
| chr17_31871058_31871230 | 0.38 |
Sik1 |
salt inducible kinase 1 |
15340 |
0.14 |
| chr8_70219826_70219982 | 0.38 |
Armc6 |
armadillo repeat containing 6 |
2386 |
0.16 |
| chr19_42601924_42602199 | 0.37 |
Loxl4 |
lysyl oxidase-like 4 |
1260 |
0.45 |
| chr1_106189902_106190053 | 0.37 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
18225 |
0.16 |
| chr1_193270900_193271248 | 0.36 |
G0s2 |
G0/G1 switch gene 2 |
2143 |
0.16 |
| chr4_54874667_54875024 | 0.36 |
Gm12479 |
predicted gene 12479 |
21717 |
0.23 |
| chr7_100908136_100908439 | 0.36 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
13597 |
0.13 |
| chr1_157012797_157012985 | 0.36 |
Gm10531 |
predicted gene 10531 |
30656 |
0.14 |
| chr2_154613231_154613406 | 0.36 |
Zfp341 |
zinc finger protein 341 |
21 |
0.6 |
| chr9_102864196_102864634 | 0.36 |
Ryk |
receptor-like tyrosine kinase |
4651 |
0.18 |
| chr4_86664611_86664778 | 0.36 |
Plin2 |
perilipin 2 |
1031 |
0.52 |
| chr9_21032221_21032372 | 0.35 |
Mir1900 |
microRNA 1900 |
32 |
0.44 |
| chr8_120244102_120244267 | 0.35 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
15728 |
0.17 |
| chr2_31135715_31135955 | 0.35 |
Fnbp1 |
formin binding protein 1 |
5967 |
0.16 |
| chr17_80061203_80061420 | 0.35 |
Hnrnpll |
heterogeneous nuclear ribonucleoprotein L-like |
824 |
0.55 |
| chr2_38379077_38379228 | 0.35 |
Gm13584 |
predicted gene 13584 |
15172 |
0.14 |
| chr8_11002584_11002773 | 0.35 |
9530052E02Rik |
RIKEN cDNA 9530052E02 gene |
5172 |
0.14 |
| chr8_83582345_83582520 | 0.34 |
Gm45823 |
predicted gene 45823 |
3093 |
0.12 |
| chr6_36280777_36280955 | 0.34 |
9330158H04Rik |
RIKEN cDNA 9330158H04 gene |
6510 |
0.29 |
| chr5_112294789_112294953 | 0.34 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
6109 |
0.12 |
| chr8_117357893_117358044 | 0.34 |
Cmip |
c-Maf inducing protein |
8798 |
0.25 |
| chr8_93959067_93959388 | 0.34 |
Gnao1 |
guanine nucleotide binding protein, alpha O |
3617 |
0.19 |
| chr15_102181335_102181499 | 0.34 |
Csad |
cysteine sulfinic acid decarboxylase |
1556 |
0.24 |
| chr16_37431973_37432137 | 0.34 |
Stxbp5l |
syntaxin binding protein 5-like |
47093 |
0.13 |
| chr4_148147807_148147968 | 0.34 |
Mir7022 |
microRNA 7022 |
891 |
0.36 |
| chr8_13038384_13038564 | 0.33 |
F10 |
coagulation factor X |
636 |
0.54 |
| chr9_65036564_65036722 | 0.33 |
Dpp8 |
dipeptidylpeptidase 8 |
3865 |
0.18 |
| chr18_69690243_69690422 | 0.33 |
Tcf4 |
transcription factor 4 |
7952 |
0.3 |
| chr13_63929441_63929609 | 0.33 |
Gm7695 |
predicted gene 7695 |
16996 |
0.18 |
| chr4_7143814_7143981 | 0.32 |
Gm11804 |
predicted gene 11804 |
104383 |
0.08 |
| chr2_44114387_44114618 | 0.32 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
49178 |
0.17 |
| chr14_79514360_79514523 | 0.32 |
Elf1 |
E74-like factor 1 |
1233 |
0.43 |
| chr7_68606375_68606542 | 0.32 |
Gm44887 |
predicted gene 44887 |
7604 |
0.22 |
| chr5_121400646_121400949 | 0.32 |
Gm43420 |
predicted gene 43420 |
1861 |
0.18 |
| chr18_80237311_80237611 | 0.31 |
Hsbp1l1 |
heat shock factor binding protein 1-like 1 |
628 |
0.57 |
| chr13_38086772_38086930 | 0.31 |
Gm27387 |
predicted gene, 27387 |
649 |
0.69 |
| chr5_142440070_142440526 | 0.31 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
23646 |
0.17 |
| chr1_51909329_51909480 | 0.31 |
Myo1b |
myosin IB |
2088 |
0.24 |
| chr15_25942169_25942359 | 0.31 |
Retreg1 |
reticulophagy regulator 1 |
415 |
0.85 |
| chr5_126954306_126954487 | 0.31 |
Gm42955 |
predicted gene 42955 |
65831 |
0.11 |
| chr6_28896982_28897268 | 0.31 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
17082 |
0.2 |
| chr8_83582597_83582758 | 0.30 |
Gm45823 |
predicted gene 45823 |
2848 |
0.13 |
| chr9_123492299_123492489 | 0.30 |
Limd1 |
LIM domains containing 1 |
11784 |
0.17 |
| chr3_145592253_145592420 | 0.30 |
Znhit6 |
zinc finger, HIT type 6 |
3532 |
0.25 |
| chr11_45877552_45877727 | 0.30 |
Clint1 |
clathrin interactor 1 |
4758 |
0.18 |
| chr11_114867107_114867285 | 0.30 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
2704 |
0.19 |
| chr2_61474961_61475340 | 0.30 |
Gm22338 |
predicted gene, 22338 |
17616 |
0.26 |
| chr5_72763116_72763280 | 0.29 |
Txk |
TXK tyrosine kinase |
10421 |
0.15 |
| chr17_5295996_5296161 | 0.29 |
Gm29050 |
predicted gene 29050 |
92685 |
0.08 |
| chr15_7070249_7070660 | 0.29 |
Lifr |
LIF receptor alpha |
20160 |
0.25 |
| chr17_27817796_27817959 | 0.29 |
Ilrun |
inflammation and lipid regulator with UBA-like and NBR1-like domains |
2575 |
0.18 |
| chr7_16047828_16047993 | 0.28 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
11 |
0.97 |
| chr6_31263686_31264023 | 0.28 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
13994 |
0.16 |
| chr8_93332780_93333003 | 0.28 |
Ces1g |
carboxylesterase 1G |
4417 |
0.17 |
| chr7_6343656_6343820 | 0.28 |
Gm3854 |
predicted gene 3854 |
21 |
0.77 |
| chr11_4661332_4661649 | 0.28 |
Ascc2 |
activating signal cointegrator 1 complex subunit 2 |
23743 |
0.12 |
| chr6_114866683_114866890 | 0.28 |
Vgll4 |
vestigial like family member 4 |
2029 |
0.33 |
| chr15_100625350_100625569 | 0.28 |
Dazap2 |
DAZ associated protein 2 |
9127 |
0.08 |
| chr18_36713491_36713783 | 0.28 |
Cd14 |
CD14 antigen |
13101 |
0.07 |
| chr19_4719341_4719664 | 0.27 |
Sptbn2 |
spectrin beta, non-erythrocytic 2 |
1577 |
0.24 |
| chr5_93250297_93250457 | 0.27 |
Ccng2 |
cyclin G2 |
16880 |
0.14 |
| chr7_127967101_127967258 | 0.27 |
Fus |
fused in sarcoma |
278 |
0.77 |
| chr2_133552164_133552315 | 0.27 |
Bmp2 |
bone morphogenetic protein 2 |
80 |
0.97 |
| chr2_93775482_93775671 | 0.27 |
Gm13818 |
predicted gene 13818 |
18994 |
0.17 |
| chr19_6991850_6992452 | 0.27 |
Dnajc4 |
DnaJ heat shock protein family (Hsp40) member C4 |
121 |
0.89 |
| chr7_112237962_112238160 | 0.27 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
12171 |
0.27 |
| chr9_115481085_115481255 | 0.27 |
Gm5921 |
predicted gene 5921 |
42589 |
0.13 |
| chr4_134488308_134488479 | 0.26 |
Gm13250 |
predicted gene 13250 |
4829 |
0.11 |
| chr11_29613290_29613486 | 0.26 |
Gm12091 |
predicted gene 12091 |
10775 |
0.13 |
| chr17_86539829_86540339 | 0.26 |
Gm10309 |
predicted gene 10309 |
34852 |
0.18 |
| chr5_146974799_146974984 | 0.26 |
Mtif3 |
mitochondrial translational initiation factor 3 |
11091 |
0.17 |
| chr6_124694855_124695031 | 0.26 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
6476 |
0.07 |
| chr5_65349405_65349570 | 0.26 |
Klb |
klotho beta |
1079 |
0.38 |
| chr14_100284919_100285099 | 0.26 |
Klf12 |
Kruppel-like factor 12 |
330 |
0.83 |
| chrX_37085164_37085323 | 0.26 |
Rpl39 |
ribosomal protein L39 |
77 |
0.94 |
| chr12_111443408_111443817 | 0.26 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
712 |
0.55 |
| chr11_7536835_7536986 | 0.26 |
Gm11986 |
predicted gene 11986 |
246536 |
0.02 |
| chr2_90503158_90503454 | 0.26 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
24056 |
0.16 |
| chr9_10340859_10341010 | 0.25 |
Gm25086 |
predicted gene, 25086 |
45778 |
0.15 |
| chr10_66933753_66933946 | 0.25 |
Gm26576 |
predicted gene, 26576 |
13547 |
0.16 |
| chr14_30906121_30906307 | 0.25 |
Itih4 |
inter alpha-trypsin inhibitor, heavy chain 4 |
7053 |
0.11 |
| chr19_7090027_7090210 | 0.25 |
AC109619.1 |
novel transcript, antisense to Macrod1 |
1697 |
0.24 |
| chr19_21914078_21914469 | 0.25 |
Ldhb-ps |
lactate dehydrogenase B, pseudogene |
23107 |
0.18 |
| chr5_117985838_117985989 | 0.25 |
Fbxo21 |
F-box protein 21 |
6589 |
0.15 |
| chr17_34586879_34587070 | 0.25 |
Notch4 |
notch 4 |
29 |
0.91 |
| chr1_164435096_164435776 | 0.25 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
8283 |
0.15 |
| chr16_77360217_77360390 | 0.25 |
Gm37466 |
predicted gene, 37466 |
21647 |
0.13 |
| chr10_93161593_93161779 | 0.25 |
Cdk17 |
cyclin-dependent kinase 17 |
796 |
0.5 |
| chrX_16815855_16816045 | 0.25 |
Maob |
monoamine oxidase B |
1325 |
0.59 |
| chr12_85785184_85785363 | 0.24 |
Flvcr2 |
feline leukemia virus subgroup C cellular receptor 2 |
10443 |
0.16 |
| chr5_35751138_35751297 | 0.24 |
Ablim2 |
actin-binding LIM protein 2 |
6663 |
0.18 |
| chr5_130256839_130256998 | 0.24 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
117 |
0.93 |
| chr16_90143819_90143995 | 0.24 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
138 |
0.96 |
| chr14_59505972_59506134 | 0.24 |
Cab39l |
calcium binding protein 39-like |
27237 |
0.17 |
| chr7_25238448_25238634 | 0.24 |
Gsk3a |
glycogen synthase kinase 3 alpha |
690 |
0.45 |
| chr11_63851241_63851480 | 0.24 |
Hmgb1-ps3 |
high mobility group box 1, pseudogene 3 |
4542 |
0.26 |
| chr10_62216958_62217109 | 0.24 |
Tspan15 |
tetraspanin 15 |
14218 |
0.15 |
| chr7_131556667_131556855 | 0.24 |
Hmx2 |
H6 homeobox 2 |
2700 |
0.19 |
| chr4_109140391_109140554 | 0.24 |
Osbpl9 |
oxysterol binding protein-like 9 |
16138 |
0.21 |
| chr17_46995065_46995238 | 0.24 |
Ubr2 |
ubiquitin protein ligase E3 component n-recognin 2 |
15328 |
0.16 |
| chr9_66795028_66795465 | 0.24 |
Aph1b |
aph1 homolog B, gamma secretase subunit |
130 |
0.93 |
| chr8_88863392_88863746 | 0.24 |
Gm45504 |
predicted gene 45504 |
495 |
0.85 |
| chr19_36834098_36834270 | 0.24 |
Tnks2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
48 |
0.98 |
| chr9_88438974_88439140 | 0.24 |
Snx14 |
sorting nexin 14 |
99 |
0.95 |
| chr7_44670891_44671049 | 0.24 |
Myh14 |
myosin, heavy polypeptide 14 |
127 |
0.92 |
| chr1_119661376_119661532 | 0.24 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
12454 |
0.16 |
| chr8_126736201_126736366 | 0.24 |
Gm45805 |
predicted gene 45805 |
22051 |
0.23 |
| chr1_21232096_21232271 | 0.23 |
Gm38224 |
predicted gene, 38224 |
2539 |
0.18 |
| chr10_125354951_125355118 | 0.23 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
12855 |
0.22 |
| chr19_53140710_53140861 | 0.23 |
Add3 |
adducin 3 (gamma) |
268 |
0.88 |
| chr8_60890550_60890715 | 0.23 |
Hpf1 |
histone PARylation factor 1 |
143 |
0.96 |
| chr11_113725569_113725747 | 0.23 |
Cdc42ep4 |
CDC42 effector protein (Rho GTPase binding) 4 |
13664 |
0.13 |
| chr16_23978554_23978740 | 0.23 |
Bcl6 |
B cell leukemia/lymphoma 6 |
6267 |
0.18 |
| chr6_88743999_88744163 | 0.23 |
Gm43999 |
predicted gene, 43999 |
1354 |
0.31 |
| chr5_122526542_122526845 | 0.23 |
Gm22965 |
predicted gene, 22965 |
19904 |
0.09 |
| chr11_28584268_28584419 | 0.23 |
Ccdc85a |
coiled-coil domain containing 85A |
19 |
0.99 |
| chr8_8994570_8994800 | 0.23 |
Gm44515 |
predicted gene 44515 |
61986 |
0.12 |
| chr9_77753424_77753607 | 0.23 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
1020 |
0.45 |
| chr11_86916528_86916698 | 0.23 |
Ypel2 |
yippee like 2 |
55411 |
0.11 |
| chr19_10073210_10073809 | 0.23 |
Fads2 |
fatty acid desaturase 2 |
2322 |
0.22 |
| chr8_25793840_25794009 | 0.23 |
Lsm1 |
LSM1 homolog, mRNA degradation associated |
8337 |
0.1 |
| chr2_12148001_12148165 | 0.22 |
Itga8 |
integrin alpha 8 |
15531 |
0.2 |
| chr14_76152803_76153117 | 0.22 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
42069 |
0.14 |
| chr15_10248990_10249545 | 0.22 |
Prlr |
prolactin receptor |
293 |
0.94 |
| chr10_59475055_59475224 | 0.22 |
Mcu |
mitochondrial calcium uniporter |
16979 |
0.18 |
| chr2_164785184_164785412 | 0.22 |
Snx21 |
sorting nexin family member 21 |
525 |
0.53 |
| chr9_69323922_69324079 | 0.22 |
Rora |
RAR-related orphan receptor alpha |
34318 |
0.15 |
| chr10_20104280_20104431 | 0.22 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
4830 |
0.24 |
| chr16_30065187_30065349 | 0.22 |
Hes1 |
hes family bHLH transcription factor 1 |
72 |
0.97 |
| chrX_12135920_12136079 | 0.22 |
Bcor |
BCL6 interacting corepressor |
7599 |
0.26 |
| chr2_59726132_59726380 | 0.22 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
16546 |
0.26 |
| chr5_144333150_144333429 | 0.22 |
Dmrt1i |
Dmrt1 interacting ncRNA |
24634 |
0.13 |
| chr19_40464469_40464658 | 0.22 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
13173 |
0.21 |
| chr4_11861590_11861758 | 0.22 |
Gm25002 |
predicted gene, 25002 |
87213 |
0.07 |
| chr6_35253418_35253775 | 0.22 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
864 |
0.54 |
| chr11_17258171_17258606 | 0.22 |
C1d |
C1D nuclear receptor co-repressor |
700 |
0.69 |
| chr4_95141392_95141560 | 0.21 |
Gm28096 |
predicted gene 28096 |
10264 |
0.16 |
| chr11_109485510_109486095 | 0.21 |
Arsg |
arylsulfatase G |
196 |
0.91 |
| chr11_115824988_115825141 | 0.21 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
1006 |
0.35 |
| chr5_43869261_43869583 | 0.21 |
Cd38 |
CD38 antigen |
562 |
0.61 |
| chr10_25595963_25596295 | 0.21 |
Gm36908 |
predicted gene, 36908 |
25434 |
0.15 |
| chr6_57506758_57506926 | 0.21 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
8765 |
0.14 |
| chr19_47337853_47338021 | 0.21 |
Sh3pxd2a |
SH3 and PX domains 2A |
23186 |
0.17 |
| chr12_28623362_28623536 | 0.21 |
Colec11 |
collectin sub-family member 11 |
72 |
0.96 |
| chr8_119417171_119417322 | 0.21 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
16878 |
0.13 |
| chr4_126181292_126181458 | 0.21 |
Thrap3 |
thyroid hormone receptor associated protein 3 |
1228 |
0.32 |
| chr8_82689441_82689600 | 0.21 |
Gm9655 |
predicted gene 9655 |
58096 |
0.13 |
| chr4_82406927_82407107 | 0.21 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
32393 |
0.21 |
| chr2_126373873_126374024 | 0.21 |
Atp8b4 |
ATPase, class I, type 8B, member 4 |
117605 |
0.05 |
| chr15_78450182_78450345 | 0.21 |
Tmprss6 |
transmembrane serine protease 6 |
5884 |
0.11 |
| chr16_33062247_33062405 | 0.21 |
Lmln |
leishmanolysin-like (metallopeptidase M8 family) |
195 |
0.9 |
| chr4_125065950_125066101 | 0.21 |
Dnali1 |
dynein, axonemal, light intermediate polypeptide 1 |
322 |
0.66 |
| chr4_128621016_128621181 | 0.21 |
Tlr12 |
toll-like receptor 12 |
2479 |
0.24 |
| chr10_81326957_81327144 | 0.21 |
Tbxa2r |
thromboxane A2 receptor |
1681 |
0.13 |
| chr13_74166717_74166944 | 0.21 |
Exoc3 |
exocyst complex component 3 |
7617 |
0.19 |
| chr11_75190453_75190617 | 0.21 |
Dph1 |
diphthamide biosynthesis 1 |
16 |
0.95 |
| chr1_73188866_73189036 | 0.21 |
Gm38106 |
predicted gene, 38106 |
20472 |
0.23 |
| chr11_58914378_58914796 | 0.21 |
Btnl10 |
butyrophilin-like 10 |
3321 |
0.09 |
| chr10_99215027_99215178 | 0.20 |
Gm34574 |
predicted gene, 34574 |
7964 |
0.12 |
| chr7_112353137_112353298 | 0.20 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
3823 |
0.33 |
| chr17_10237978_10238400 | 0.20 |
Qk |
quaking |
24615 |
0.24 |
| chr5_8961694_8961940 | 0.20 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
3265 |
0.17 |
| chr6_13871418_13871584 | 0.20 |
2610001J05Rik |
RIKEN cDNA 2610001J05 gene |
17 |
0.5 |
| chr5_150195920_150196236 | 0.20 |
Fry |
FRY microtubule binding protein |
388 |
0.88 |
| chr11_19920515_19920734 | 0.20 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
3751 |
0.33 |
| chr17_12415417_12415751 | 0.20 |
Plg |
plasminogen |
36925 |
0.12 |
| chr17_85037271_85037422 | 0.20 |
Slc3a1 |
solute carrier family 3, member 1 |
8970 |
0.19 |
| chr3_115635601_115635754 | 0.20 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
79395 |
0.09 |
| chr2_34154899_34155111 | 0.20 |
Gm38389 |
predicted gene, 38389 |
43561 |
0.15 |
| chr15_76722928_76723137 | 0.20 |
C030006K11Rik |
RIKEN cDNA C030006K11 gene |
353 |
0.62 |
| chr19_40630689_40630866 | 0.20 |
Gm8783 |
predicted pseudogene 8783 |
8317 |
0.12 |
| chr17_12402018_12402192 | 0.20 |
Plg |
plasminogen |
23446 |
0.15 |
| chr1_52113713_52114086 | 0.20 |
Gm31812 |
predicted gene, 31812 |
148 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.1 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.2 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.2 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.2 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:2001198 | regulation of dendritic cell differentiation(GO:2001198) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.0 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0043307 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.0 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.0 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0071332 | response to fructose(GO:0009750) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.2 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0034791 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |