Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
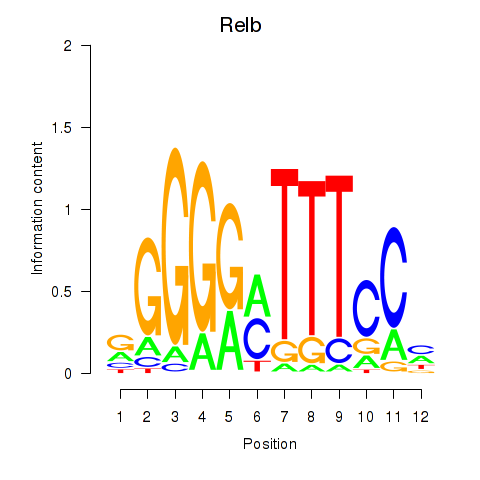
Results for Relb
Z-value: 0.89
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSMUSG00000002983.10 | avian reticuloendotheliosis viral (v-rel) oncogene related B |
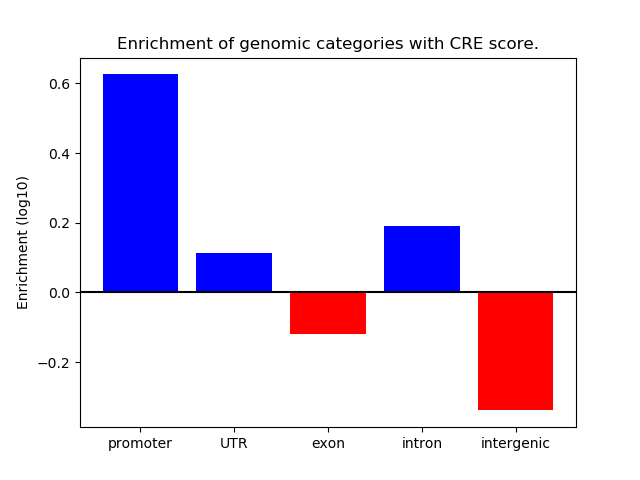
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_19631131_19631299 | Relb | 1777 | 0.175563 | -0.73 | 9.8e-02 | Click! |
| chr7_19619136_19619293 | Relb | 9220 | 0.084860 | -0.70 | 1.2e-01 | Click! |
| chr7_19628765_19628929 | Relb | 63 | 0.938451 | 0.46 | 3.6e-01 | Click! |
| chr7_19628515_19628680 | Relb | 124 | 0.914468 | 0.44 | 3.8e-01 | Click! |
| chr7_19629111_19629282 | Relb | 52 | 0.941131 | 0.15 | 7.8e-01 | Click! |
Activity of the Relb motif across conditions
Conditions sorted by the z-value of the Relb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
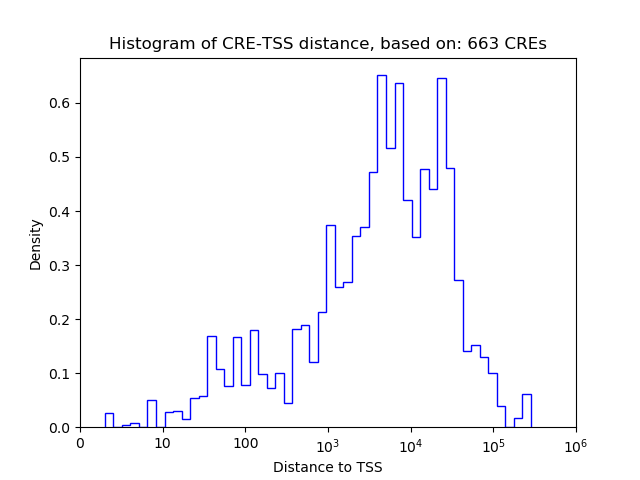
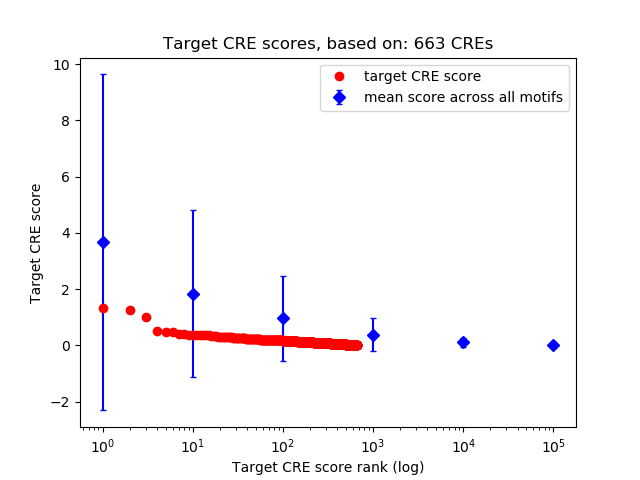
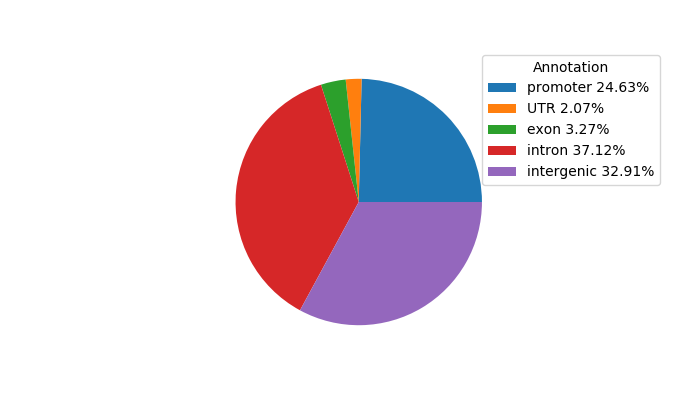
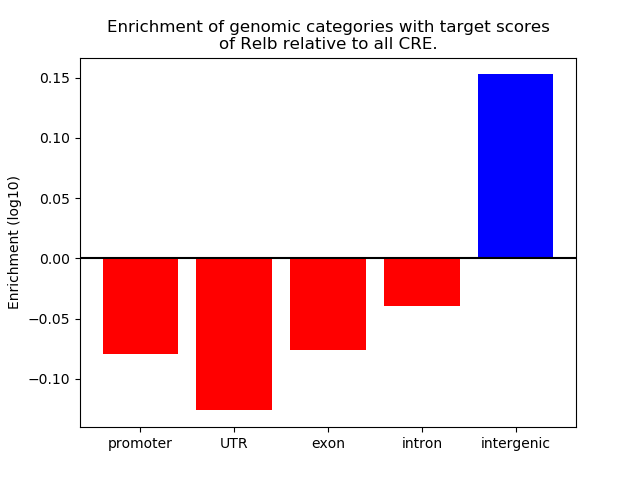
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_141628129_141628296 | 1.33 |
Mir7063 |
microRNA 7063 |
7511 |
0.13 |
| chr17_31828262_31828413 | 1.24 |
Sik1 |
salt inducible kinase 1 |
23238 |
0.14 |
| chr6_125527760_125527927 | 1.01 |
Vwf |
Von Willebrand factor |
18931 |
0.15 |
| chr8_11373743_11373894 | 0.52 |
Col4a2 |
collagen, type IV, alpha 2 |
31090 |
0.13 |
| chr10_38970068_38970231 | 0.48 |
Lama4 |
laminin, alpha 4 |
4477 |
0.27 |
| chr13_93989307_93989458 | 0.48 |
Gm47216 |
predicted gene, 47216 |
2386 |
0.28 |
| chr7_46723990_46724178 | 0.41 |
Saa3 |
serum amyloid A 3 |
8384 |
0.09 |
| chr6_99375099_99375287 | 0.40 |
Gm20705 |
predicted gene 20705 |
13306 |
0.23 |
| chr4_43492855_43493028 | 0.38 |
Ccdc107 |
coiled-coil domain containing 107 |
41 |
0.58 |
| chr12_104247702_104247874 | 0.38 |
Serpina3h |
serine (or cysteine) peptidase inhibitor, clade A, member 3H |
117 |
0.93 |
| chr2_169790587_169791240 | 0.37 |
Tshz2 |
teashirt zinc finger family member 2 |
93980 |
0.08 |
| chr12_8386824_8386996 | 0.37 |
Gm48075 |
predicted gene, 48075 |
7299 |
0.18 |
| chr2_129340331_129340510 | 0.37 |
Gm14039 |
predicted gene 14039 |
22439 |
0.09 |
| chr8_127196588_127196779 | 0.37 |
Pard3 |
par-3 family cell polarity regulator |
25279 |
0.24 |
| chr12_79600452_79600623 | 0.36 |
Rad51b |
RAD51 paralog B |
273184 |
0.01 |
| chr6_115996180_115996399 | 0.34 |
Plxnd1 |
plexin D1 |
1284 |
0.4 |
| chr17_45557476_45557627 | 0.33 |
Nfkbie |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
1812 |
0.18 |
| chr18_75405710_75405888 | 0.33 |
Smad7 |
SMAD family member 7 |
30885 |
0.18 |
| chr1_182758644_182758936 | 0.31 |
Susd4 |
sushi domain containing 4 |
5070 |
0.21 |
| chr9_5308705_5308876 | 0.30 |
Casp4 |
caspase 4, apoptosis-related cysteine peptidase |
38 |
0.98 |
| chr6_94996352_94996560 | 0.29 |
4930511E03Rik |
RIKEN cDNA 4930511E03 gene |
52622 |
0.13 |
| chr1_57989000_57989391 | 0.29 |
Sgo2a |
shugoshin 2A |
6776 |
0.19 |
| chr6_3384983_3385179 | 0.29 |
Samd9l |
sterile alpha motif domain containing 9-like |
75 |
0.97 |
| chr17_87365352_87365503 | 0.29 |
0610012D04Rik |
RIKEN cDNA 0610012D04 gene |
965 |
0.48 |
| chr15_66814050_66814231 | 0.29 |
Sla |
src-like adaptor |
1547 |
0.4 |
| chr2_35658400_35658598 | 0.29 |
Dab2ip |
disabled 2 interacting protein |
2851 |
0.31 |
| chr6_121303535_121303719 | 0.28 |
Slc6a13 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
2845 |
0.19 |
| chr2_25534308_25534469 | 0.28 |
Traf2 |
TNF receptor-associated factor 2 |
4752 |
0.08 |
| chr3_105803714_105803865 | 0.28 |
Rap1a |
RAS-related protein 1a |
2453 |
0.17 |
| chr8_11394936_11395099 | 0.27 |
Col4a2 |
collagen, type IV, alpha 2 |
9891 |
0.16 |
| chr10_127962398_127962784 | 0.27 |
Gm47949 |
predicted gene, 47949 |
1035 |
0.33 |
| chr3_97232474_97232625 | 0.26 |
Bcl9 |
B cell CLL/lymphoma 9 |
3703 |
0.21 |
| chr8_48944740_48944906 | 0.26 |
Gm25830 |
predicted gene, 25830 |
13658 |
0.25 |
| chr16_50753691_50753842 | 0.26 |
Dubr |
Dppa2 upstream binding RNA |
20993 |
0.17 |
| chr11_114867640_114867800 | 0.25 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
3228 |
0.17 |
| chr11_16906555_16906817 | 0.25 |
Egfr |
epidermal growth factor receptor |
1501 |
0.41 |
| chr9_113749434_113749589 | 0.24 |
Clasp2 |
CLIP associating protein 2 |
7779 |
0.2 |
| chr11_98459290_98459441 | 0.24 |
Grb7 |
growth factor receptor bound protein 7 |
5576 |
0.1 |
| chr2_93214308_93214479 | 0.24 |
Tspan18 |
tetraspanin 18 |
13393 |
0.2 |
| chr2_85172170_85172321 | 0.24 |
Gm13713 |
predicted gene 13713 |
11467 |
0.11 |
| chr5_75377802_75378010 | 0.24 |
Gm22084 |
predicted gene, 22084 |
5761 |
0.2 |
| chr18_60803553_60803864 | 0.23 |
Cd74 |
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
140 |
0.94 |
| chr1_155063987_155064316 | 0.23 |
Gm29282 |
predicted gene 29282 |
6972 |
0.17 |
| chr14_20970523_20970838 | 0.23 |
Vcl |
vinculin |
41282 |
0.14 |
| chr5_143055727_143055894 | 0.22 |
Gm43378 |
predicted gene 43378 |
5767 |
0.15 |
| chr1_4622515_4622679 | 0.22 |
Gm7369 |
predicted gene 7369 |
12126 |
0.15 |
| chr7_141429982_141430161 | 0.22 |
Cend1 |
cell cycle exit and neuronal differentiation 1 |
580 |
0.43 |
| chr5_91961314_91961670 | 0.22 |
Thap6 |
THAP domain containing 6 |
897 |
0.35 |
| chr12_55492611_55492778 | 0.22 |
Nfkbia |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
47 |
0.97 |
| chr1_40221808_40222119 | 0.22 |
Il1r1 |
interleukin 1 receptor, type I |
3117 |
0.26 |
| chr17_24204987_24205163 | 0.22 |
Tbc1d24 |
TBC1 domain family, member 24 |
407 |
0.66 |
| chr3_95635932_95636096 | 0.21 |
E330034L11Rik |
RIKEN cDNA E330034L11 gene |
3290 |
0.12 |
| chr8_11221370_11221555 | 0.21 |
Col4a1 |
collagen, type IV, alpha 1 |
4193 |
0.21 |
| chr7_25120549_25120733 | 0.21 |
Pou2f2 |
POU domain, class 2, transcription factor 2 |
217 |
0.89 |
| chr3_149326826_149327020 | 0.21 |
Gm30382 |
predicted gene, 30382 |
3049 |
0.25 |
| chr17_83277496_83277647 | 0.21 |
Gm24240 |
predicted gene, 24240 |
7600 |
0.2 |
| chr13_64179357_64179534 | 0.21 |
Habp4 |
hyaluronic acid binding protein 4 |
5058 |
0.13 |
| chr5_123607373_123607580 | 0.21 |
Clip1 |
CAP-GLY domain containing linker protein 1 |
3666 |
0.14 |
| chr7_16830530_16830865 | 0.21 |
Strn4 |
striatin, calmodulin binding protein 4 |
402 |
0.73 |
| chr19_41347409_41347564 | 0.20 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
37610 |
0.17 |
| chr7_68709256_68709407 | 0.20 |
Gm44692 |
predicted gene 44692 |
17136 |
0.21 |
| chr14_79512054_79512205 | 0.20 |
Elf1 |
E74-like factor 1 |
3545 |
0.2 |
| chr10_4135364_4135517 | 0.20 |
Gm25515 |
predicted gene, 25515 |
31127 |
0.16 |
| chr8_124634513_124634671 | 0.20 |
Capn9 |
calpain 9 |
21152 |
0.14 |
| chr10_112953941_112954181 | 0.20 |
Gm47519 |
predicted gene, 47519 |
6127 |
0.18 |
| chr3_10310720_10310871 | 0.20 |
Gm38364 |
predicted gene, 38364 |
1747 |
0.2 |
| chr15_31367525_31367697 | 0.20 |
Ankrd33b |
ankyrin repeat domain 33B |
43 |
0.93 |
| chr5_86885878_86886051 | 0.20 |
Ugt2b34 |
UDP glucuronosyltransferase 2 family, polypeptide B34 |
20973 |
0.1 |
| chr19_44900544_44900723 | 0.20 |
Slf2 |
SMC5-SMC6 complex localization factor 2 |
30518 |
0.11 |
| chr12_104262958_104263126 | 0.20 |
Serpina3i |
serine (or cysteine) peptidase inhibitor, clade A, member 3I |
80 |
0.94 |
| chr12_86612676_86612827 | 0.20 |
Gm8504 |
predicted gene 8504 |
37822 |
0.13 |
| chr2_70559621_70559796 | 0.20 |
Gad1 |
glutamate decarboxylase 1 |
2334 |
0.22 |
| chr4_123412152_123412346 | 0.20 |
Macf1 |
microtubule-actin crosslinking factor 1 |
21 |
0.97 |
| chr2_50964157_50964313 | 0.20 |
Gm13498 |
predicted gene 13498 |
54551 |
0.16 |
| chr15_85611891_85612067 | 0.19 |
Wnt7b |
wingless-type MMTV integration site family, member 7B |
29506 |
0.12 |
| chr1_133493490_133493663 | 0.19 |
Gm8596 |
predicted gene 8596 |
35903 |
0.14 |
| chr10_4107762_4107973 | 0.19 |
Gm25515 |
predicted gene, 25515 |
3554 |
0.27 |
| chr10_19087108_19087313 | 0.19 |
1700124M09Rik |
RIKEN cDNA 1700124M09 gene |
32 |
0.97 |
| chr10_80050367_80050530 | 0.19 |
Gpx4 |
glutathione peroxidase 4 |
3040 |
0.12 |
| chr19_46484022_46484320 | 0.19 |
Sufu |
SUFU negative regulator of hedgehog signaling |
1074 |
0.46 |
| chr8_120738772_120739041 | 0.19 |
Irf8 |
interferon regulatory factor 8 |
267 |
0.89 |
| chr12_13177714_13177865 | 0.19 |
Gm24208 |
predicted gene, 24208 |
14026 |
0.16 |
| chr11_72953535_72953727 | 0.18 |
Gm44467 |
predicted gene, 44467 |
524 |
0.7 |
| chr11_59804844_59804999 | 0.18 |
Flcn |
folliculin |
4667 |
0.11 |
| chr19_37239252_37239415 | 0.18 |
Gm25268 |
predicted gene, 25268 |
5849 |
0.15 |
| chr8_46739715_46739866 | 0.18 |
Irf2 |
interferon regulatory factor 2 |
6 |
0.8 |
| chr2_60283319_60283522 | 0.18 |
Cd302 |
CD302 antigen |
998 |
0.5 |
| chr4_40873404_40873555 | 0.18 |
B4galt1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
19474 |
0.1 |
| chr17_31827276_31827455 | 0.18 |
Sik1 |
salt inducible kinase 1 |
24210 |
0.14 |
| chr7_19867840_19868009 | 0.18 |
Gm44659 |
predicted gene 44659 |
3514 |
0.09 |
| chr4_47251261_47251412 | 0.18 |
Col15a1 |
collagen, type XV, alpha 1 |
7199 |
0.22 |
| chr11_77792590_77792910 | 0.18 |
Gm10277 |
predicted gene 10277 |
5003 |
0.16 |
| chr15_22791886_22792091 | 0.18 |
Hnrnpa1l2-ps2 |
heterogeneous nuclear ribonucleoprotein A1-like 2, pseudogene 2 |
77158 |
0.12 |
| chr19_10599348_10599499 | 0.18 |
Tkfc |
triokinase, FMN cyclase |
4835 |
0.1 |
| chr10_62231949_62232100 | 0.17 |
Tspan15 |
tetraspanin 15 |
773 |
0.6 |
| chr9_72660282_72660433 | 0.17 |
Nedd4 |
neural precursor cell expressed, developmentally down-regulated 4 |
1989 |
0.18 |
| chr2_152343770_152343958 | 0.17 |
Trib3 |
tribbles pseudokinase 3 |
106 |
0.92 |
| chr4_104814139_104814345 | 0.17 |
C8b |
complement component 8, beta polypeptide |
15034 |
0.21 |
| chr2_129380876_129381038 | 0.17 |
Spcs2-ps |
signal peptidase complex subunit 2, pseudogene |
1561 |
0.27 |
| chr4_150787040_150787400 | 0.17 |
Gm13049 |
predicted gene 13049 |
38513 |
0.13 |
| chr2_122158376_122158599 | 0.17 |
Trim69 |
tripartite motif-containing 69 |
2213 |
0.19 |
| chr6_31075878_31076320 | 0.17 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
11505 |
0.11 |
| chr7_80130219_80130425 | 0.17 |
Idh2 |
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
14930 |
0.1 |
| chr5_149045840_149046001 | 0.17 |
Hmgb1 |
high mobility group box 1 |
4527 |
0.13 |
| chr4_128989568_128989760 | 0.17 |
Ak2 |
adenylate kinase 2 |
2294 |
0.25 |
| chr3_97972968_97973119 | 0.17 |
Notch2 |
notch 2 |
40484 |
0.11 |
| chr14_29987413_29987605 | 0.16 |
Mir6947 |
microRNA 6947 |
1379 |
0.24 |
| chr11_16773276_16773438 | 0.16 |
Egfr |
epidermal growth factor receptor |
21127 |
0.18 |
| chr19_32298377_32298611 | 0.16 |
Sgms1 |
sphingomyelin synthase 1 |
8915 |
0.24 |
| chr4_53187279_53187439 | 0.16 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
27464 |
0.15 |
| chr6_113668529_113668873 | 0.16 |
Irak2 |
interleukin-1 receptor-associated kinase 2 |
149 |
0.89 |
| chr7_110689414_110689588 | 0.16 |
Gm21123 |
predicted gene, 21123 |
10233 |
0.15 |
| chr3_135307876_135308027 | 0.16 |
Slc9b2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
206 |
0.92 |
| chr11_100719386_100719554 | 0.16 |
Hspb9 |
heat shock protein, alpha-crystallin-related, B9 |
5548 |
0.1 |
| chr17_3989752_3989920 | 0.16 |
4930470H14Rik |
RIKEN cDNA 4930470H14 gene |
93159 |
0.09 |
| chrX_20862192_20862360 | 0.16 |
Mir5617 |
microRNA 5617 |
906 |
0.41 |
| chr2_147782596_147782759 | 0.16 |
A530006G24Rik |
RIKEN cDNA A530006G24 gene |
71984 |
0.1 |
| chr14_100284919_100285099 | 0.15 |
Klf12 |
Kruppel-like factor 12 |
330 |
0.83 |
| chr6_136938692_136939081 | 0.15 |
Arhgdib |
Rho, GDP dissociation inhibitor (GDI) beta |
705 |
0.59 |
| chr9_98675729_98675890 | 0.15 |
Pisrt1 |
polled intersex syndrome regulated transcript 1 |
30457 |
0.13 |
| chr10_94093554_94093705 | 0.15 |
Gm18391 |
predicted gene, 18391 |
4511 |
0.15 |
| chr10_91080633_91080784 | 0.15 |
Apaf1 |
apoptotic peptidase activating factor 1 |
1721 |
0.28 |
| chr7_16312800_16312989 | 0.15 |
Bbc3 |
BCL2 binding component 3 |
623 |
0.6 |
| chr1_136859525_136859810 | 0.15 |
Gm17781 |
predicted gene, 17781 |
60708 |
0.09 |
| chr7_3290355_3290515 | 0.15 |
Myadm |
myeloid-associated differentiation marker |
99 |
0.9 |
| chr12_69365915_69366084 | 0.15 |
Gm18113 |
predicted gene, 18113 |
4024 |
0.11 |
| chr11_34086328_34086662 | 0.15 |
Lcp2 |
lymphocyte cytosolic protein 2 |
509 |
0.8 |
| chr10_127147265_127147440 | 0.15 |
Gm23664 |
predicted gene, 23664 |
8735 |
0.08 |
| chr17_79068246_79068411 | 0.15 |
Qpct |
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
8674 |
0.17 |
| chr6_113238509_113238719 | 0.15 |
Mtmr14 |
myotubularin related protein 14 |
717 |
0.56 |
| chr16_29882571_29882722 | 0.15 |
Gm32679 |
predicted gene, 32679 |
47918 |
0.12 |
| chr12_91696351_91696512 | 0.15 |
Gm8378 |
predicted gene 8378 |
10318 |
0.16 |
| chr14_63899614_63899769 | 0.14 |
Pinx1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
78 |
0.98 |
| chr18_61360632_61360830 | 0.14 |
Gm25301 |
predicted gene, 25301 |
37082 |
0.11 |
| chr7_79311533_79311809 | 0.14 |
Gm39041 |
predicted gene, 39041 |
12646 |
0.14 |
| chr16_31066805_31066992 | 0.14 |
Xxylt1 |
xyloside xylosyltransferase 1 |
955 |
0.57 |
| chr14_101652712_101652891 | 0.14 |
Uchl3 |
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
1166 |
0.44 |
| chr2_158302944_158303106 | 0.14 |
Lbp |
lipopolysaccharide binding protein |
3468 |
0.15 |
| chr11_5734457_5734620 | 0.14 |
Urgcp |
upregulator of cell proliferation |
6608 |
0.13 |
| chr12_86904302_86904504 | 0.14 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
19605 |
0.16 |
| chr11_54956028_54956193 | 0.14 |
Tnip1 |
TNFAIP3 interacting protein 1 |
6 |
0.97 |
| chr10_3718557_3718760 | 0.14 |
Plekhg1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
21706 |
0.25 |
| chr8_128488839_128489004 | 0.14 |
Nrp1 |
neuropilin 1 |
129524 |
0.05 |
| chr7_98535989_98536157 | 0.14 |
A630091E08Rik |
RIKEN cDNA A630091E08 gene |
26917 |
0.12 |
| chr18_61331637_61332092 | 0.13 |
Pde6a |
phosphodiesterase 6A, cGMP-specific, rod, alpha |
56841 |
0.08 |
| chr2_10128611_10128912 | 0.13 |
Itih2 |
inter-alpha trypsin inhibitor, heavy chain 2 |
1902 |
0.23 |
| chr5_90891009_90891200 | 0.13 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
137 |
0.93 |
| chr11_61608212_61608363 | 0.13 |
Epn2 |
epsin 2 |
28600 |
0.13 |
| chr19_47330818_47330996 | 0.13 |
Sh3pxd2a |
SH3 and PX domains 2A |
16156 |
0.18 |
| chr12_84193897_84194204 | 0.13 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
35 |
0.95 |
| chr5_88796583_88796899 | 0.13 |
Gm42912 |
predicted gene 42912 |
2845 |
0.21 |
| chr1_23268164_23268335 | 0.13 |
Mir30a |
microRNA 30a |
4020 |
0.14 |
| chr5_110980028_110980206 | 0.13 |
Gm42778 |
predicted gene 42778 |
1452 |
0.34 |
| chr17_34627479_34627653 | 0.13 |
Ppt2 |
palmitoyl-protein thioesterase 2 |
41 |
0.69 |
| chr9_66276951_66277383 | 0.13 |
Dapk2 |
death-associated protein kinase 2 |
8793 |
0.23 |
| chr12_26356269_26356452 | 0.13 |
4930549C15Rik |
RIKEN cDNA 4930549C15 gene |
9643 |
0.19 |
| chr14_27101523_27101762 | 0.13 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
13257 |
0.19 |
| chr10_59441733_59442104 | 0.13 |
Oit3 |
oncoprotein induced transcript 3 |
140 |
0.96 |
| chr10_77434735_77434942 | 0.13 |
Adarb1 |
adenosine deaminase, RNA-specific, B1 |
16568 |
0.16 |
| chr11_101965679_101965853 | 0.13 |
Sost |
sclerostin |
1249 |
0.27 |
| chr17_74630898_74631071 | 0.13 |
Birc6 |
baculoviral IAP repeat-containing 6 |
4738 |
0.22 |
| chr14_76698781_76698988 | 0.12 |
1700108F19Rik |
RIKEN cDNA 1700108F19 gene |
17130 |
0.2 |
| chr1_133863573_133863878 | 0.12 |
Gm28441 |
predicted gene 28441 |
28536 |
0.11 |
| chr15_36361442_36361595 | 0.12 |
Gm33936 |
predicted gene, 33936 |
5589 |
0.14 |
| chr4_138065715_138066098 | 0.12 |
Eif4g3 |
eukaryotic translation initiation factor 4 gamma, 3 |
61154 |
0.1 |
| chr17_74331435_74331594 | 0.12 |
Gm9351 |
predicted gene 9351 |
1015 |
0.34 |
| chr9_70328368_70328519 | 0.12 |
Mir5626 |
microRNA 5626 |
77310 |
0.08 |
| chr15_82294106_82294257 | 0.12 |
Wbp2nl |
WBP2 N-terminal like |
4773 |
0.09 |
| chr14_118591597_118591757 | 0.12 |
Mir6391 |
microRNA 6391 |
795 |
0.61 |
| chr1_170589945_170590114 | 0.12 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
168 |
0.96 |
| chr2_118513077_118513244 | 0.12 |
Gm13983 |
predicted gene 13983 |
28983 |
0.12 |
| chr5_129021137_129021430 | 0.12 |
Ran |
RAN, member RAS oncogene family |
1011 |
0.56 |
| chr8_125557731_125557884 | 0.12 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
11896 |
0.23 |
| chr16_23204539_23205077 | 0.12 |
Gm15746 |
predicted gene 15746 |
6687 |
0.1 |
| chr15_102671840_102672003 | 0.12 |
Mir688 |
microRNA 688 |
55 |
0.85 |
| chr17_29395434_29395615 | 0.12 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
19023 |
0.11 |
| chr1_195012643_195012824 | 0.12 |
9630010A21Rik |
RIKEN cDNA 9630010A21 gene |
1013 |
0.32 |
| chr6_97457292_97457492 | 0.12 |
Frmd4b |
FERM domain containing 4B |
1894 |
0.36 |
| chr5_123257174_123257325 | 0.12 |
Rpl35a-ps6 |
ribosomal protein L35A, pseudogene 6 |
1436 |
0.26 |
| chr16_23060324_23060755 | 0.12 |
Kng1 |
kininogen 1 |
2146 |
0.13 |
| chr11_4974491_4974717 | 0.12 |
Ap1b1 |
adaptor protein complex AP-1, beta 1 subunit |
12220 |
0.13 |
| chr4_139611543_139611704 | 0.12 |
Gm21969 |
predicted gene 21969 |
8437 |
0.14 |
| chr2_70870934_70871085 | 0.12 |
Gm13632 |
predicted gene 13632 |
6820 |
0.2 |
| chr11_97406666_97406821 | 0.11 |
Arhgap23 |
Rho GTPase activating protein 23 |
8790 |
0.15 |
| chr5_113207341_113207492 | 0.11 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
13820 |
0.13 |
| chr9_53390887_53391256 | 0.11 |
Poglut3 |
protein O-glucosyltransferase 3 |
454 |
0.78 |
| chr10_93919382_93919541 | 0.11 |
Gm48126 |
predicted gene, 48126 |
371 |
0.79 |
| chr16_4399765_4400133 | 0.11 |
Adcy9 |
adenylate cyclase 9 |
19638 |
0.19 |
| chr5_137493763_137493914 | 0.11 |
Epo |
erythropoietin |
8022 |
0.07 |
| chr13_107676889_107677059 | 0.11 |
1700006H21Rik |
RIKEN cDNA 1700006H21 gene |
10423 |
0.23 |
| chr4_133703497_133703648 | 0.11 |
Mir7227 |
microRNA 7227 |
13536 |
0.12 |
| chr19_55916323_55916507 | 0.11 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
18106 |
0.24 |
| chr2_122147524_122147696 | 0.11 |
B2m |
beta-2 microglobulin |
76 |
0.95 |
| chr19_55063941_55064146 | 0.11 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
35408 |
0.17 |
| chr19_29138834_29139228 | 0.11 |
Mir101b |
microRNA 101b |
3752 |
0.18 |
| chr13_31180346_31180513 | 0.11 |
Gm11372 |
predicted gene 11372 |
18657 |
0.21 |
| chr5_135118666_135118877 | 0.11 |
Gm43500 |
predicted gene 43500 |
4522 |
0.11 |
| chr19_26951406_26951563 | 0.11 |
Gm815 |
predicted gene 815 |
65559 |
0.11 |
| chr1_36064998_36065160 | 0.11 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
3321 |
0.19 |
| chr9_66510150_66510491 | 0.11 |
Fbxl22 |
F-box and leucine-rich repeat protein 22 |
4289 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.4 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:1903935 | response to sodium arsenite(GO:1903935) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.0 | 0.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.2 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.0 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.2 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0034920 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0003933 | GTP cyclohydrolase activity(GO:0003933) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |