Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
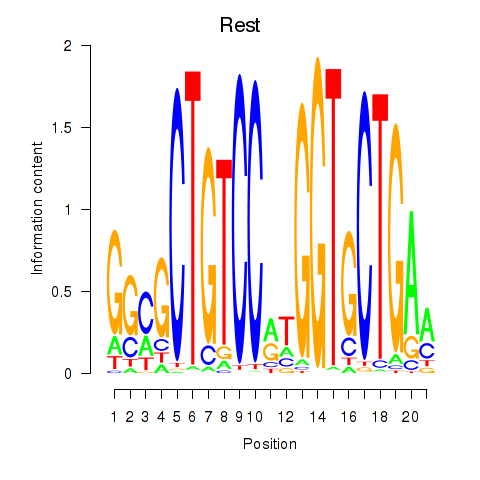
Results for Rest
Z-value: 2.88
Transcription factors associated with Rest
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rest
|
ENSMUSG00000029249.9 | RE1-silencing transcription factor |
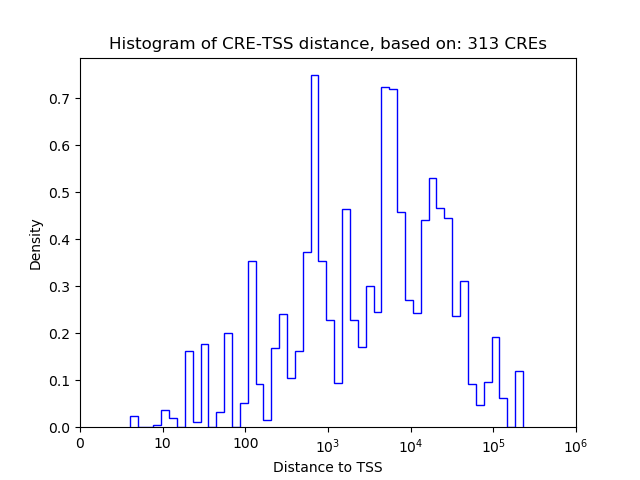
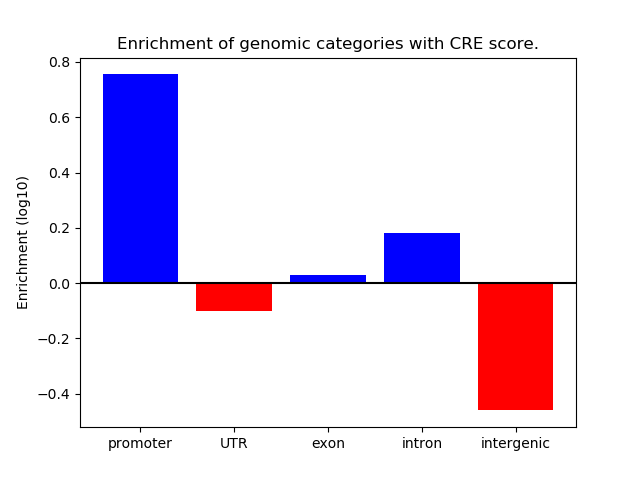
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_77267056_77267224 | Rest | 944 | 0.451623 | 0.96 | 1.8e-03 | Click! |
| chr5_77266781_77267041 | Rest | 715 | 0.567690 | 0.78 | 6.4e-02 | Click! |
| chr5_77265048_77265375 | Rest | 280 | 0.865173 | -0.69 | 1.3e-01 | Click! |
| chr5_77266106_77266414 | Rest | 64 | 0.959682 | -0.64 | 1.7e-01 | Click! |
| chr5_77265452_77265845 | Rest | 157 | 0.931221 | -0.48 | 3.3e-01 | Click! |
Activity of the Rest motif across conditions
Conditions sorted by the z-value of the Rest motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
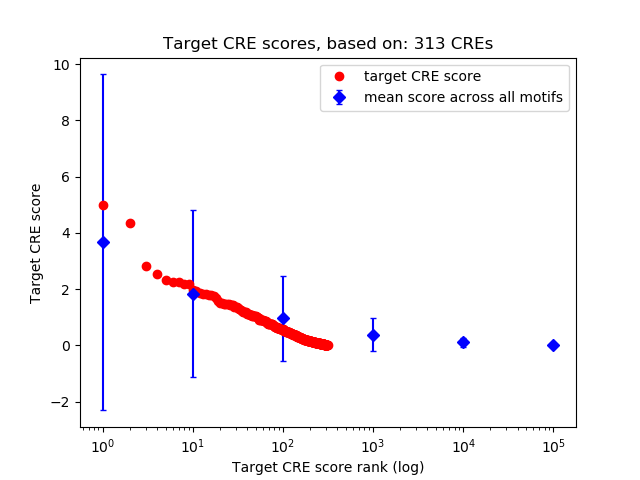
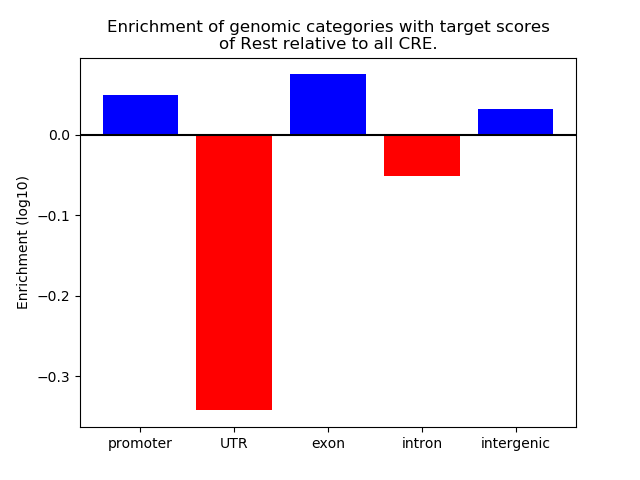
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_75173089_75173918 | 4.99 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr18_25668171_25668329 | 4.35 |
0710001A04Rik |
RIKEN cDNA 0710001A04 gene |
45520 |
0.16 |
| chr4_131115371_131115541 | 2.81 |
A930031H19Rik |
RIKEN cDNA A930031H19 gene |
104982 |
0.06 |
| chr15_74076059_74076222 | 2.53 |
Gm7935 |
predicted pseudogene 7935 |
5371 |
0.24 |
| chr15_84829493_84829651 | 2.32 |
Phf21b |
PHD finger protein 21B |
25538 |
0.14 |
| chr15_79791641_79791792 | 2.27 |
Nptxr |
neuronal pentraxin receptor |
735 |
0.49 |
| chr4_140283535_140283688 | 2.26 |
Igsf21 |
immunoglobulin superfamily, member 21 |
36827 |
0.14 |
| chr9_40801417_40801568 | 2.19 |
Hspa8 |
heat shock protein 8 |
31 |
0.94 |
| chr19_10403213_10403386 | 2.18 |
Syt7 |
synaptotagmin VII |
719 |
0.62 |
| chr1_173361798_173361949 | 1.97 |
Cadm3 |
cell adhesion molecule 3 |
5760 |
0.17 |
| chr1_75277299_75277450 | 1.94 |
Resp18 |
regulated endocrine-specific protein 18 |
910 |
0.34 |
| chr7_4996598_4996749 | 1.87 |
Zfp579 |
zinc finger protein 579 |
515 |
0.52 |
| chr8_14072651_14072815 | 1.84 |
Gm45520 |
predicted gene 45520 |
4915 |
0.16 |
| chr6_55108463_55108627 | 1.81 |
Crhr2 |
corticotropin releasing hormone receptor 2 |
7972 |
0.19 |
| chr6_113962934_113963105 | 1.81 |
Gm15082 |
predicted gene 15082 |
25280 |
0.14 |
| chr4_151604592_151604765 | 1.79 |
4930589P08Rik |
RIKEN cDNA 4930589P08 gene |
228208 |
0.02 |
| chr9_121791324_121791483 | 1.77 |
Hhatl |
hedgehog acyltransferase-like |
57 |
0.94 |
| chr19_6427254_6427405 | 1.68 |
Nrxn2 |
neurexin II |
594 |
0.57 |
| chr2_158570467_158570618 | 1.58 |
Arhgap40 |
Rho GTPase activating protein 40 |
28805 |
0.1 |
| chr11_118584913_118585075 | 1.52 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
15084 |
0.22 |
| chr9_106889455_106889671 | 1.49 |
Manf |
mesencephalic astrocyte-derived neurotrophic factor |
720 |
0.47 |
| chr9_21950442_21950600 | 1.48 |
Swsap1 |
SWIM type zinc finger 7 associated protein 1 |
5234 |
0.09 |
| chr7_138084146_138084302 | 1.47 |
Gm25798 |
predicted gene, 25798 |
17315 |
0.23 |
| chr9_74535088_74535239 | 1.47 |
Gm28622 |
predicted gene 28622 |
26215 |
0.2 |
| chr4_107019507_107019668 | 1.47 |
Gm12786 |
predicted gene 12786 |
2912 |
0.21 |
| chr11_83421415_83421571 | 1.44 |
Gas2l2 |
growth arrest-specific 2 like 2 |
7962 |
0.11 |
| chr7_30484338_30484496 | 1.44 |
Nphs1 |
nephrosis 1, nephrin |
498 |
0.41 |
| chr19_38558935_38559114 | 1.42 |
Plce1 |
phospholipase C, epsilon 1 |
24344 |
0.22 |
| chr6_89614766_89614917 | 1.38 |
Chchd6 |
coiled-coil-helix-coiled-coil-helix domain containing 6 |
19189 |
0.17 |
| chr15_84853852_84854164 | 1.35 |
Phf21b |
PHD finger protein 21B |
1102 |
0.45 |
| chr15_89150169_89150320 | 1.35 |
Mapk11 |
mitogen-activated protein kinase 11 |
616 |
0.52 |
| chr5_33438295_33438455 | 1.34 |
Gm43851 |
predicted gene 43851 |
911 |
0.54 |
| chr7_44770135_44770325 | 1.28 |
Vrk3 |
vaccinia related kinase 3 |
13244 |
0.08 |
| chr12_104850467_104850964 | 1.26 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
6656 |
0.19 |
| chr9_119389058_119389253 | 1.22 |
Xylb |
xylulokinase homolog (H. influenzae) |
10964 |
0.11 |
| chr4_45844423_45844581 | 1.20 |
Stra6l |
STRA6-like |
4162 |
0.18 |
| chr10_45546948_45547107 | 1.20 |
Gm27731 |
predicted gene, 27731 |
9783 |
0.18 |
| chr13_117604223_117604403 | 1.19 |
Hcn1 |
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
1833 |
0.49 |
| chr7_30434353_30434522 | 1.19 |
Gm30646 |
predicted gene, 30646 |
1893 |
0.12 |
| chr4_62257049_62257210 | 1.12 |
Slc31a2 |
solute carrier family 31, member 2 |
5433 |
0.16 |
| chr3_19694558_19694716 | 1.11 |
Crh |
corticotropin releasing hormone |
759 |
0.59 |
| chr5_24357551_24357718 | 1.10 |
Kcnh2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
6030 |
0.1 |
| chr1_74066477_74066680 | 1.07 |
Tns1 |
tensin 1 |
24773 |
0.17 |
| chr11_83417193_83417347 | 1.07 |
Rasl10b |
RAS-like, family 10, member B |
7198 |
0.11 |
| chr16_18087231_18087692 | 1.06 |
Prodh |
proline dehydrogenase |
1561 |
0.29 |
| chr15_72568672_72568828 | 1.06 |
Gm49460 |
predicted gene, 49460 |
19533 |
0.2 |
| chr9_122006687_122006862 | 1.06 |
Gm47117 |
predicted gene, 47117 |
5366 |
0.11 |
| chr9_20857229_20857389 | 1.05 |
Shfl |
shiftless antiviral inhibitor of ribosomal frameshifting |
11333 |
0.08 |
| chr12_102550629_102550788 | 1.03 |
Chga |
chromogranin A |
4261 |
0.19 |
| chr4_115728832_115728993 | 1.03 |
Efcab14 |
EF-hand calcium binding domain 14 |
8832 |
0.14 |
| chr9_122116996_122117412 | 1.02 |
Snrk |
SNF related kinase |
62 |
0.95 |
| chr11_103920773_103920924 | 1.00 |
Nsf |
N-ethylmaleimide sensitive fusion protein |
5303 |
0.22 |
| chr8_123838618_123838800 | 0.95 |
2810455O05Rik |
RIKEN cDNA 2810455O05 gene |
2076 |
0.17 |
| chr3_94374766_94374934 | 0.95 |
Rorc |
RAR-related orphan receptor gamma |
2056 |
0.12 |
| chr11_115374569_115374733 | 0.91 |
Hid1 |
HID1 domain containing |
6895 |
0.09 |
| chr19_44692476_44692627 | 0.91 |
Gm26644 |
predicted gene, 26644 |
2724 |
0.24 |
| chr2_109675977_109676138 | 0.91 |
Bdnf |
brain derived neurotrophic factor |
160 |
0.83 |
| chr19_47016602_47016771 | 0.90 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
1533 |
0.25 |
| chr6_90513392_90513725 | 0.90 |
Gm44236 |
predicted gene, 44236 |
256 |
0.88 |
| chr1_127496856_127497018 | 0.90 |
Tmem163 |
transmembrane protein 163 |
3407 |
0.32 |
| chr2_180104544_180104879 | 0.87 |
Hrh3 |
histamine receptor H3 |
223 |
0.88 |
| chr5_35531711_35531875 | 0.87 |
Cpz |
carboxypeptidase Z |
6095 |
0.15 |
| chr9_60982764_60983106 | 0.86 |
Gm5122 |
predicted gene 5122 |
30546 |
0.14 |
| chr11_74619258_74619412 | 0.86 |
Ccdc92b |
coiled-coil domain containing 92B |
270 |
0.9 |
| chr3_88220963_88221116 | 0.85 |
Gm3764 |
predicted gene 3764 |
1625 |
0.17 |
| chr1_85080827_85081001 | 0.81 |
Gm18342 |
predicted gene, 18342 |
678 |
0.51 |
| chr7_4620106_4620257 | 0.80 |
Gm44877 |
predicted gene 44877 |
6797 |
0.07 |
| chr6_85927460_85927847 | 0.78 |
Nat8b-ps |
N-acetyltransferase 8B, pseudogene |
5726 |
0.08 |
| chr7_44593894_44594057 | 0.77 |
Kcnc3 |
potassium voltage gated channel, Shaw-related subfamily, member 3 |
20 |
0.93 |
| chr2_178277324_178277480 | 0.77 |
Phactr3 |
phosphatase and actin regulator 3 |
50154 |
0.15 |
| chr1_85500331_85500508 | 0.77 |
Gm2427 |
predicted gene 2427 |
671 |
0.57 |
| chr1_85179698_85179873 | 0.76 |
Gm6264 |
predicted gene 6264 |
18922 |
0.09 |
| chr8_71483947_71484113 | 0.76 |
Ano8 |
anoctamin 8 |
19 |
0.94 |
| chr4_140358223_140358381 | 0.76 |
Gm13016 |
predicted gene 13016 |
22328 |
0.14 |
| chr2_91110132_91110333 | 0.76 |
Mybpc3 |
myosin binding protein C, cardiac |
7912 |
0.11 |
| chr9_9230047_9230379 | 0.75 |
Gm16833 |
predicted gene, 16833 |
6075 |
0.24 |
| chr17_29492005_29492199 | 0.73 |
Pim1 |
proviral integration site 1 |
94 |
0.61 |
| chr9_111139563_111139885 | 0.72 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
20049 |
0.15 |
| chr19_42326023_42326184 | 0.68 |
Crtac1 |
cartilage acidic protein 1 |
38149 |
0.11 |
| chr11_81486379_81486540 | 0.67 |
1700071K01Rik |
RIKEN cDNA 1700071K01 gene |
87014 |
0.09 |
| chr2_180756003_180756165 | 0.66 |
Slc17a9 |
solute carrier family 17, member 9 |
17810 |
0.1 |
| chr18_54799055_54799220 | 0.66 |
Gm33732 |
predicted gene, 33732 |
24652 |
0.16 |
| chr12_8099948_8100109 | 0.66 |
Apob |
apolipoprotein B |
87669 |
0.08 |
| chr14_32467734_32467885 | 0.65 |
Chat |
choline acetyltransferase |
1820 |
0.24 |
| chr6_121473169_121473328 | 0.63 |
Iqsec3 |
IQ motif and Sec7 domain 3 |
375 |
0.88 |
| chr6_113428446_113428672 | 0.63 |
Cidec |
cell death-inducing DFFA-like effector c |
6054 |
0.07 |
| chr12_108994751_108994937 | 0.63 |
Gm18405 |
predicted gene, 18405 |
3193 |
0.19 |
| chr13_78171316_78171488 | 0.62 |
3110006O06Rik |
RIKEN cDNA 3110006O06 gene |
228 |
0.9 |
| chr13_55445821_55446370 | 0.62 |
Grk6 |
G protein-coupled receptor kinase 6 |
634 |
0.5 |
| chr6_83927539_83927712 | 0.61 |
Zfp638 |
zinc finger protein 638 |
1230 |
0.35 |
| chr17_69439256_69439434 | 0.60 |
Akain1 |
A kinase (PRKA) anchor inhibitor 1 |
19 |
0.51 |
| chr6_72634309_72634791 | 0.58 |
Gm15401 |
predicted gene 15401 |
1703 |
0.17 |
| chr5_141985119_141985282 | 0.58 |
Sdk1 |
sidekick cell adhesion molecule 1 |
128413 |
0.06 |
| chr7_57592274_57592432 | 0.58 |
Gabrb3 |
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
823 |
0.67 |
| chr15_26309788_26309969 | 0.58 |
Marchf11 |
membrane associated ring-CH-type finger 11 |
814 |
0.77 |
| chr11_31888267_31888568 | 0.57 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
15142 |
0.2 |
| chr9_107486954_107487108 | 0.57 |
Cacna2d2 |
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
18573 |
0.08 |
| chr6_128469552_128469734 | 0.57 |
Gm8719 |
predicted pseudogene 8719 |
3745 |
0.1 |
| chr6_33249470_33249649 | 0.57 |
Exoc4 |
exocyst complex component 4 |
423 |
0.81 |
| chr8_112128950_112129112 | 0.56 |
Gm22827 |
predicted gene, 22827 |
66526 |
0.1 |
| chr11_117872991_117873321 | 0.55 |
Tha1 |
threonine aldolase 1 |
303 |
0.79 |
| chr2_26208213_26208385 | 0.55 |
Lhx3 |
LIM homeobox protein 3 |
10 |
0.95 |
| chr12_112141395_112141548 | 0.53 |
Kif26a |
kinesin family member 26A |
4737 |
0.15 |
| chr6_108430619_108430854 | 0.51 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
12859 |
0.2 |
| chr11_43902830_43903004 | 0.51 |
Adra1b |
adrenergic receptor, alpha 1b |
1707 |
0.46 |
| chr14_47486187_47486558 | 0.51 |
Gm48986 |
predicted gene, 48986 |
5196 |
0.14 |
| chr5_45529813_45529965 | 0.50 |
Gm42521 |
predicted gene 42521 |
1183 |
0.35 |
| chr5_110558497_110558658 | 0.50 |
Galnt9 |
polypeptide N-acetylgalactosaminyltransferase 9 |
14222 |
0.14 |
| chr4_46992957_46993118 | 0.49 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
1164 |
0.45 |
| chr2_120050008_120050173 | 0.49 |
Pla2g4b |
phospholipase A2, group IVB (cytosolic) |
9120 |
0.11 |
| chr9_110993538_110993895 | 0.48 |
Rtp3 |
receptor transporter protein 3 |
3133 |
0.12 |
| chr17_30389798_30389963 | 0.48 |
Gm6402 |
predicted pseudogene 6402 |
6153 |
0.19 |
| chr1_60628564_60628727 | 0.47 |
Gm23762 |
predicted gene, 23762 |
14842 |
0.14 |
| chr15_78066618_78066776 | 0.47 |
A730060N03Rik |
RIKEN cDNA A730060N03 gene |
53009 |
0.09 |
| chr18_15434023_15434197 | 0.47 |
Aqp4 |
aquaporin 4 |
23128 |
0.2 |
| chr7_30346746_30346906 | 0.46 |
4930479H17Rik |
RIKEN cDNA 4930479H17 gene |
6495 |
0.07 |
| chr2_43613684_43613852 | 0.46 |
Kynu |
kynureninase |
14153 |
0.29 |
| chr9_43698520_43698693 | 0.45 |
Gm5364 |
predicted gene 5364 |
15553 |
0.14 |
| chr17_15077876_15078027 | 0.45 |
Ermard |
ER membrane associated RNA degradation |
12336 |
0.15 |
| chr3_94914590_94914772 | 0.45 |
Gm26279 |
predicted gene, 26279 |
5742 |
0.11 |
| chr10_80748733_80748893 | 0.45 |
Dot1l |
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
6393 |
0.09 |
| chr2_70559621_70559796 | 0.44 |
Gad1 |
glutamate decarboxylase 1 |
2334 |
0.22 |
| chr11_116108287_116108624 | 0.42 |
Trim47 |
tripartite motif-containing 47 |
336 |
0.77 |
| chr10_79718092_79718250 | 0.42 |
Hcn2 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
1537 |
0.18 |
| chr5_139014946_139015101 | 0.41 |
Prkar1b |
protein kinase, cAMP dependent regulatory, type I beta |
8787 |
0.18 |
| chr16_5146310_5146979 | 0.41 |
Sec14l5 |
SEC14-like lipid binding 5 |
465 |
0.73 |
| chr1_88194822_88195116 | 0.39 |
Gm15376 |
predicted gene 15376 |
3068 |
0.09 |
| chr9_29146016_29146172 | 0.39 |
Ntm |
neurotrimin |
36312 |
0.22 |
| chr13_95782265_95782699 | 0.39 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
18085 |
0.18 |
| chr2_65619900_65620061 | 0.39 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
791 |
0.72 |
| chr16_32914274_32914432 | 0.38 |
Lrch3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
211 |
0.91 |
| chr2_30065686_30066045 | 0.38 |
Set |
SET nuclear oncogene |
591 |
0.59 |
| chr7_99699987_99700199 | 0.37 |
Slco2b1 |
solute carrier organic anion transporter family, member 2b1 |
4284 |
0.12 |
| chr11_75046982_75047192 | 0.37 |
Gm12333 |
predicted gene 12333 |
7739 |
0.13 |
| chr14_122513626_122513788 | 0.36 |
Gm18143 |
predicted gene, 18143 |
122 |
0.94 |
| chr1_175665774_175665943 | 0.36 |
Chml |
choroideremia-like |
22495 |
0.13 |
| chr1_67235018_67235169 | 0.36 |
Gm15668 |
predicted gene 15668 |
14107 |
0.22 |
| chr4_117893494_117893672 | 0.36 |
Ipo13 |
importin 13 |
1531 |
0.19 |
| chr12_9166308_9166471 | 0.35 |
Ttc32 |
tetratricopeptide repeat domain 32 |
136361 |
0.04 |
| chr5_146238766_146238947 | 0.35 |
Cdk8 |
cyclin-dependent kinase 8 |
7471 |
0.12 |
| chr15_28024715_28025200 | 0.35 |
Trio |
triple functional domain (PTPRF interacting) |
3 |
0.98 |
| chr2_79454254_79454412 | 0.35 |
Neurod1 |
neurogenic differentiation 1 |
2418 |
0.28 |
| chr4_117850329_117850493 | 0.34 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
1041 |
0.37 |
| chr1_162548031_162548373 | 0.32 |
Eef1aknmt |
EEF1A lysine and N-terminal methyltransferase |
54 |
0.97 |
| chr4_120126163_120126326 | 0.31 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
31755 |
0.17 |
| chr8_69088523_69088826 | 0.31 |
Atp6v1b2 |
ATPase, H+ transporting, lysosomal V1 subunit B2 |
28 |
0.91 |
| chr11_119007787_119007988 | 0.31 |
Cbx2 |
chromobox 2 |
15075 |
0.15 |
| chr1_85586586_85586850 | 0.30 |
Sp110 |
Sp110 nuclear body protein |
2440 |
0.15 |
| chr11_115258288_115258439 | 0.29 |
Gm25837 |
predicted gene, 25837 |
5085 |
0.11 |
| chr7_142387074_142387268 | 0.29 |
Ctsd |
cathepsin D |
648 |
0.53 |
| chr11_69914167_69914731 | 0.29 |
Gps2 |
G protein pathway suppressor 2 |
257 |
0.73 |
| chr13_54371670_54371821 | 0.28 |
Cplx2 |
complexin 2 |
396 |
0.83 |
| chr4_63254251_63254424 | 0.28 |
Mir455 |
microRNA 455 |
2514 |
0.25 |
| chr19_16447855_16448017 | 0.28 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
8636 |
0.18 |
| chr14_20093617_20093768 | 0.27 |
Saysd1 |
SAYSVFN motif domain containing 1 |
10509 |
0.17 |
| chr11_69934461_69934654 | 0.27 |
Ybx2 |
Y box protein 2 |
1239 |
0.18 |
| chr6_113881955_113882116 | 0.26 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
9335 |
0.18 |
| chr9_48603402_48603655 | 0.26 |
Nnmt |
nicotinamide N-methyltransferase |
1549 |
0.47 |
| chr5_123395114_123395623 | 0.26 |
Gm15747 |
predicted gene 15747 |
304 |
0.6 |
| chr11_79427788_79427962 | 0.25 |
Nf1 |
neurofibromin 1 |
20278 |
0.13 |
| chr4_12906721_12906911 | 0.25 |
Triqk |
triple QxxK/R motif containing |
22 |
0.99 |
| chr19_31890378_31890565 | 0.24 |
A1cf |
APOBEC1 complementation factor |
21690 |
0.19 |
| chr11_104114823_104114983 | 0.24 |
Crhr1 |
corticotropin releasing hormone receptor 1 |
17952 |
0.21 |
| chr16_45742916_45743108 | 0.24 |
Abhd10 |
abhydrolase domain containing 10 |
57 |
0.96 |
| chr9_56508889_56509217 | 0.23 |
Gm47175 |
predicted gene, 47175 |
2392 |
0.25 |
| chr16_30266715_30266896 | 0.23 |
Cpn2 |
carboxypeptidase N, polypeptide 2 |
694 |
0.61 |
| chr5_24416500_24416681 | 0.23 |
Asic3 |
acid-sensing (proton-gated) ion channel 3 |
13 |
0.93 |
| chr11_72608017_72608168 | 0.22 |
Ube2g1 |
ubiquitin-conjugating enzyme E2G 1 |
773 |
0.57 |
| chr1_75179893_75180252 | 0.22 |
Abcb6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
214 |
0.81 |
| chr8_119876752_119876924 | 0.22 |
Klhl36 |
kelch-like 36 |
14566 |
0.15 |
| chr6_121144204_121144363 | 0.21 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
13284 |
0.14 |
| chr2_31641363_31641698 | 0.21 |
Gm13425 |
predicted gene 13425 |
941 |
0.35 |
| chr13_113612458_113612853 | 0.21 |
Snx18 |
sorting nexin 18 |
986 |
0.54 |
| chr11_110101538_110102129 | 0.20 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
5855 |
0.27 |
| chr6_120463585_120463736 | 0.20 |
Il17ra |
interleukin 17 receptor A |
413 |
0.8 |
| chr1_188118233_188118393 | 0.20 |
Gm38315 |
predicted gene, 38315 |
42394 |
0.18 |
| chr2_157913212_157913385 | 0.20 |
Vstm2l |
V-set and transmembrane domain containing 2-like |
1355 |
0.44 |
| chr2_93906445_93906632 | 0.20 |
Gm1335 |
predicted gene 1335 |
34340 |
0.11 |
| chr19_23160573_23160812 | 0.20 |
Mir1192 |
microRNA 1192 |
11261 |
0.16 |
| chr10_87528215_87528468 | 0.19 |
Pah |
phenylalanine hydroxylase |
6314 |
0.21 |
| chr9_121311967_121312154 | 0.19 |
Trak1 |
trafficking protein, kinesin binding 1 |
2178 |
0.24 |
| chr8_11478332_11478510 | 0.18 |
Rab20 |
RAB20, member RAS oncogene family |
289 |
0.63 |
| chr3_89764370_89764528 | 0.18 |
Chrnb2 |
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) |
159 |
0.92 |
| chr3_107279311_107279673 | 0.18 |
Lamtor5 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
493 |
0.74 |
| chr13_96962673_96962863 | 0.18 |
Gm48597 |
predicted gene, 48597 |
11854 |
0.16 |
| chr12_104025874_104026200 | 0.18 |
Gm37496 |
predicted gene, 37496 |
1610 |
0.23 |
| chr2_91921277_91921473 | 0.18 |
Gm10805 |
predicted gene 10805 |
447 |
0.72 |
| chr11_16860070_16860314 | 0.17 |
Egfr |
epidermal growth factor receptor |
17958 |
0.18 |
| chr7_45198547_45198718 | 0.17 |
Ccdc155 |
coiled-coil domain containing 155 |
6260 |
0.06 |
| chr4_137676216_137676391 | 0.17 |
Rap1gap |
Rap1 GTPase-activating protein |
4450 |
0.19 |
| chr17_72837306_72837660 | 0.16 |
Ypel5 |
yippee like 5 |
53 |
0.98 |
| chr9_48675246_48675425 | 0.16 |
Nnmt |
nicotinamide N-methyltransferase |
70182 |
0.11 |
| chr5_100673558_100673901 | 0.16 |
Coq2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
427 |
0.78 |
| chr18_61665532_61665824 | 0.16 |
Carmn |
cardiac mesoderm enhancer-associated non-coding RNA |
141 |
0.92 |
| chr3_28264177_28264328 | 0.16 |
Tnik |
TRAF2 and NCK interacting kinase |
609 |
0.76 |
| chr10_81419902_81420248 | 0.16 |
Mir1191b |
microRNA 1191b |
3778 |
0.08 |
| chr17_85877554_85877736 | 0.15 |
Gm30117 |
predicted gene, 30117 |
50934 |
0.14 |
| chr17_24876191_24876410 | 0.15 |
Igfals |
insulin-like growth factor binding protein, acid labile subunit |
1356 |
0.21 |
| chrX_75874057_75874225 | 0.15 |
Pls3 |
plastin 3 (T-isoform) |
433 |
0.81 |
| chr1_92039344_92039495 | 0.14 |
Gm37036 |
predicted gene, 37036 |
17322 |
0.19 |
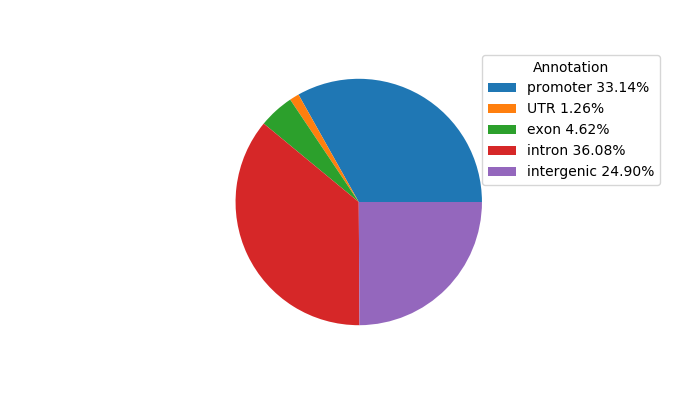
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 1.7 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.3 | 4.3 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.2 | 1.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.7 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.2 | 0.6 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 0.8 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.2 | 0.8 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 0.7 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 1.0 | GO:0098598 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.1 | 0.4 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.2 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.3 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 0.5 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.3 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.5 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.1 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.5 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.2 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.1 | 0.2 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.7 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.1 | GO:2000591 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.0 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.0 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.0 | 0.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0060061 | regulation of collateral sprouting in absence of injury(GO:0048696) Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.0 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 0.7 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.4 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.7 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 1.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.0 | GO:0018585 | fluorene oxygenase activity(GO:0018585) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.0 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |