Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
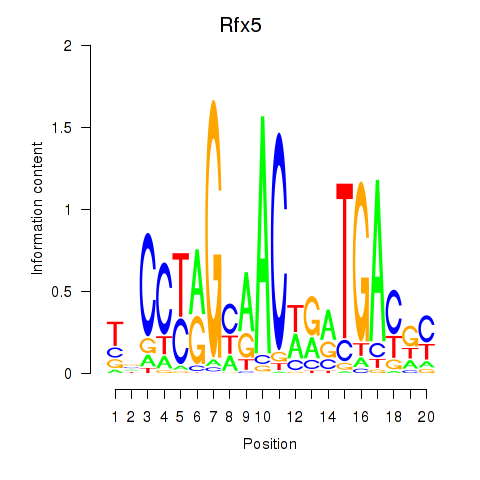
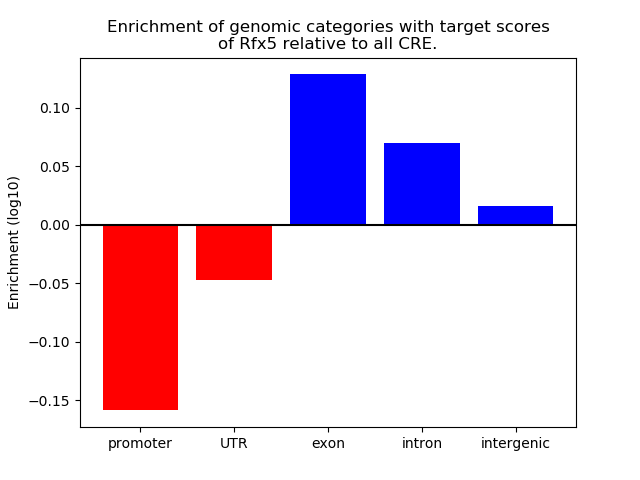
Results for Rfx5
Z-value: 0.79
Transcription factors associated with Rfx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx5
|
ENSMUSG00000005774.6 | regulatory factor X, 5 (influences HLA class II expression) |
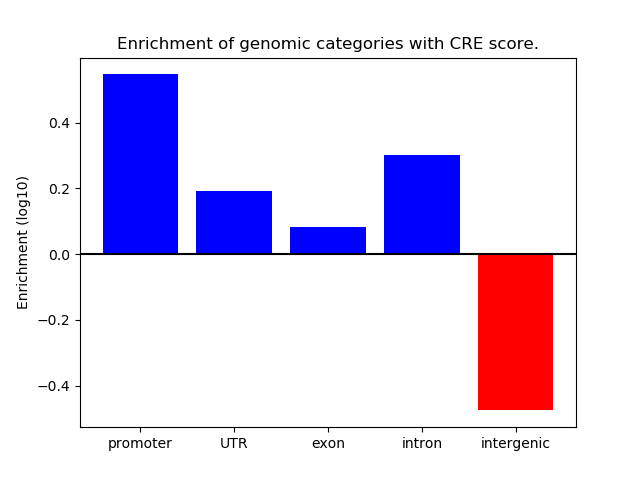
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_94953385_94953872 | Rfx5 | 447 | 0.655877 | -0.78 | 6.7e-02 | Click! |
| chr3_94945756_94945907 | Rfx5 | 8244 | 0.088078 | 0.74 | 9.4e-02 | Click! |
| chr3_94954465_94954628 | Rfx5 | 19 | 0.946414 | -0.71 | 1.1e-01 | Click! |
| chr3_94943842_94944021 | Rfx5 | 10144 | 0.085767 | 0.53 | 2.8e-01 | Click! |
| chr3_94950096_94950252 | Rfx5 | 3901 | 0.105431 | -0.45 | 3.8e-01 | Click! |
Activity of the Rfx5 motif across conditions
Conditions sorted by the z-value of the Rfx5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_16868019_16868198 | 0.66 |
Egfr |
epidermal growth factor receptor |
10042 |
0.2 |
| chr13_23299091_23299367 | 0.63 |
Gm11333 |
predicted gene 11333 |
1180 |
0.26 |
| chr2_58769900_58770051 | 0.56 |
Upp2 |
uridine phosphorylase 2 |
4650 |
0.24 |
| chr7_63922331_63923024 | 0.56 |
Klf13 |
Kruppel-like factor 13 |
2193 |
0.22 |
| chr11_16781411_16781603 | 0.56 |
Egfr |
epidermal growth factor receptor |
29277 |
0.16 |
| chrX_156479377_156479543 | 0.55 |
Gm23404 |
predicted gene, 23404 |
23365 |
0.24 |
| chr1_67132947_67133268 | 0.50 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
10081 |
0.24 |
| chr13_108367613_108367783 | 0.47 |
Depdc1b |
DEP domain containing 1B |
10272 |
0.21 |
| chr17_37190817_37190981 | 0.42 |
Ubd |
ubiquitin D |
2993 |
0.09 |
| chr15_57710929_57711112 | 0.41 |
Gm16006 |
predicted gene 16006 |
2093 |
0.32 |
| chr13_107642057_107642316 | 0.39 |
Gm32090 |
predicted gene, 32090 |
29337 |
0.18 |
| chr1_41559831_41559986 | 0.38 |
Gm28634 |
predicted gene 28634 |
30365 |
0.26 |
| chr17_46053621_46054045 | 0.38 |
Vegfa |
vascular endothelial growth factor A |
21464 |
0.12 |
| chr17_34153027_34153195 | 0.37 |
H2-DMb1 |
histocompatibility 2, class II, locus Mb1 |
39 |
0.92 |
| chr14_65050597_65050753 | 0.35 |
Extl3 |
exostosin-like glycosyltransferase 3 |
47430 |
0.13 |
| chr15_3398579_3398730 | 0.31 |
Ghr |
growth hormone receptor |
72990 |
0.11 |
| chr7_29303739_29303929 | 0.30 |
Dpf1 |
D4, zinc and double PHD fingers family 1 |
117 |
0.93 |
| chr11_4195737_4195910 | 0.30 |
Tbc1d10a |
TBC1 domain family, member 10a |
9001 |
0.1 |
| chr19_38015663_38015814 | 0.29 |
Myof |
myoferlin |
27613 |
0.14 |
| chr4_76360115_76360401 | 0.28 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
16015 |
0.23 |
| chr2_73491256_73491419 | 0.27 |
Wipf1 |
WAS/WASL interacting protein family, member 1 |
4868 |
0.19 |
| chr11_16744767_16744918 | 0.27 |
Egfr |
epidermal growth factor receptor |
7361 |
0.2 |
| chr11_16790428_16790580 | 0.26 |
Egfr |
epidermal growth factor receptor |
38274 |
0.14 |
| chr8_77045181_77045332 | 0.26 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
82757 |
0.09 |
| chr11_16893244_16893395 | 0.26 |
Egfr |
epidermal growth factor receptor |
11866 |
0.19 |
| chr11_16837434_16837735 | 0.26 |
Egfros |
epidermal growth factor receptor, opposite strand |
6882 |
0.22 |
| chr14_77586069_77586220 | 0.25 |
Enox1 |
ecto-NOX disulfide-thiol exchanger 1 |
13288 |
0.18 |
| chr3_50077080_50077240 | 0.25 |
Gm37826 |
predicted gene, 37826 |
47491 |
0.16 |
| chr15_59065322_59065483 | 0.25 |
Mtss1 |
MTSS I-BAR domain containing 1 |
9938 |
0.23 |
| chr6_142528529_142528737 | 0.25 |
Ldhb |
lactate dehydrogenase B |
20676 |
0.17 |
| chr15_63155793_63155965 | 0.24 |
Gm49013 |
predicted gene, 49013 |
9646 |
0.23 |
| chr2_27770776_27770927 | 0.24 |
Rxra |
retinoid X receptor alpha |
30650 |
0.2 |
| chr19_8757228_8757608 | 0.24 |
Nxf1 |
nuclear RNA export factor 1 |
268 |
0.68 |
| chr5_125483749_125483900 | 0.24 |
Gm27551 |
predicted gene, 27551 |
4447 |
0.15 |
| chr3_63317524_63317675 | 0.24 |
Mme |
membrane metallo endopeptidase |
17439 |
0.25 |
| chr17_34347674_34347859 | 0.24 |
H2-Ea |
histocompatibility 2, class II antigen E alpha |
3121 |
0.11 |
| chr2_172934925_172935100 | 0.23 |
Bmp7 |
bone morphogenetic protein 7 |
5080 |
0.21 |
| chr16_37938812_37938975 | 0.22 |
Gpr156 |
G protein-coupled receptor 156 |
22397 |
0.15 |
| chr4_54733994_54734173 | 0.22 |
Gm12477 |
predicted gene 12477 |
63891 |
0.11 |
| chr11_16817302_16817617 | 0.22 |
Egfros |
epidermal growth factor receptor, opposite strand |
13243 |
0.21 |
| chr1_59201583_59201922 | 0.22 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
17374 |
0.14 |
| chr16_78322780_78322958 | 0.21 |
Cxadr |
coxsackie virus and adenovirus receptor |
2338 |
0.25 |
| chr1_52022235_52022386 | 0.21 |
Stat4 |
signal transducer and activator of transcription 4 |
13684 |
0.16 |
| chr3_89142602_89142893 | 0.21 |
Pklr |
pyruvate kinase liver and red blood cell |
6124 |
0.07 |
| chr15_58957352_58957568 | 0.20 |
Mtss1 |
MTSS I-BAR domain containing 1 |
620 |
0.66 |
| chr3_27562067_27562227 | 0.20 |
Gm43344 |
predicted gene 43344 |
15506 |
0.22 |
| chr10_50623725_50623913 | 0.20 |
Ascc3 |
activating signal cointegrator 1 complex subunit 3 |
18623 |
0.22 |
| chr1_91446944_91447095 | 0.20 |
Per2 |
period circadian clock 2 |
2846 |
0.16 |
| chr17_34257684_34257866 | 0.20 |
H2-Ab1 |
histocompatibility 2, class II antigen A, beta 1 |
86 |
0.92 |
| chr7_135747212_135747378 | 0.20 |
Gm36431 |
predicted gene, 36431 |
4619 |
0.2 |
| chr17_34304881_34305040 | 0.19 |
Gm20513 |
predicted gene 20513 |
760 |
0.33 |
| chr17_83057976_83058141 | 0.19 |
Gm29052 |
predicted gene 29052 |
10847 |
0.21 |
| chr6_51578310_51578498 | 0.19 |
Snx10 |
sorting nexin 10 |
33838 |
0.16 |
| chr17_34305717_34305873 | 0.19 |
H2-Eb1 |
histocompatibility 2, class II antigen E beta |
72 |
0.49 |
| chr10_80679534_80679874 | 0.19 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
1592 |
0.19 |
| chr6_136553862_136554030 | 0.19 |
Atf7ip |
activating transcription factor 7 interacting protein |
58 |
0.97 |
| chr4_108119092_108119243 | 0.19 |
Scp2 |
sterol carrier protein 2, liver |
635 |
0.66 |
| chr14_64098292_64098443 | 0.18 |
Prss51 |
protease, serine 51 |
4641 |
0.13 |
| chr5_122313082_122313277 | 0.18 |
Gm15842 |
predicted gene 15842 |
10757 |
0.1 |
| chr5_53994743_53994917 | 0.18 |
Stim2 |
stromal interaction molecule 2 |
3669 |
0.3 |
| chr13_59893097_59893271 | 0.18 |
Gm19866 |
predicted gene, 19866 |
13935 |
0.13 |
| chr16_43346186_43346337 | 0.18 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
16787 |
0.16 |
| chr6_108638094_108638414 | 0.18 |
Gm17055 |
predicted gene 17055 |
18663 |
0.14 |
| chr1_91444586_91444906 | 0.18 |
Per2 |
period circadian clock 2 |
5119 |
0.13 |
| chr11_61223499_61223666 | 0.18 |
Aldh3a1 |
aldehyde dehydrogenase family 3, subfamily A1 |
14837 |
0.13 |
| chr3_142619782_142619948 | 0.18 |
Gbp2 |
guanylate binding protein 2 |
737 |
0.55 |
| chr18_60803553_60803864 | 0.18 |
Cd74 |
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
140 |
0.94 |
| chr1_150873368_150873519 | 0.18 |
Hmcn1 |
hemicentin 1 |
119608 |
0.05 |
| chr5_118649434_118649712 | 0.18 |
Gm43274 |
predicted gene 43274 |
5200 |
0.18 |
| chr5_33605422_33605612 | 0.18 |
Fam53a |
family with sequence similarity 53, member A |
23395 |
0.11 |
| chr1_92937865_92938056 | 0.18 |
Capn10 |
calpain 10 |
2499 |
0.15 |
| chr2_45463913_45464105 | 0.18 |
Gm13479 |
predicted gene 13479 |
106744 |
0.07 |
| chr10_80668723_80668916 | 0.18 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
3157 |
0.11 |
| chr1_13467289_13467464 | 0.17 |
Gm7593 |
predicted gene 7593 |
59721 |
0.1 |
| chr17_34079687_34079853 | 0.17 |
H2-Pb |
histocompatibility 2, P region beta locus |
3603 |
0.06 |
| chr11_120806039_120806735 | 0.17 |
Fasn |
fatty acid synthase |
2218 |
0.14 |
| chr1_45672893_45673066 | 0.17 |
Gm23216 |
predicted gene, 23216 |
12583 |
0.19 |
| chr17_84647222_84647423 | 0.17 |
Dync2li1 |
dynein cytoplasmic 2 light intermediate chain 1 |
1953 |
0.24 |
| chr16_33932119_33932270 | 0.17 |
Itgb5 |
integrin beta 5 |
9708 |
0.17 |
| chr8_69074605_69074774 | 0.17 |
Slc18a1 |
solute carrier family 18 (vesicular monoamine), member 1 |
819 |
0.58 |
| chr8_105086546_105086842 | 0.17 |
Ces3b |
carboxylesterase 3B |
1925 |
0.2 |
| chr19_10067201_10067465 | 0.16 |
Fads2 |
fatty acid desaturase 2 |
3854 |
0.16 |
| chr11_48843419_48843570 | 0.16 |
Trim7 |
tripartite motif-containing 7 |
4408 |
0.1 |
| chr16_35471313_35471717 | 0.16 |
Pdia5 |
protein disulfide isomerase associated 5 |
19074 |
0.14 |
| chr9_74704204_74704355 | 0.16 |
Gm27233 |
predicted gene 27233 |
4983 |
0.27 |
| chr3_9610870_9611024 | 0.16 |
Zfp704 |
zinc finger protein 704 |
862 |
0.67 |
| chr1_154118196_154118387 | 0.16 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
340 |
0.89 |
| chr7_84129291_84129727 | 0.15 |
Abhd17c |
abhydrolase domain containing 17C |
18306 |
0.14 |
| chr11_120817967_120818292 | 0.15 |
Fasn |
fatty acid synthase |
2674 |
0.13 |
| chr14_118008711_118008863 | 0.15 |
Dct |
dopachrome tautomerase |
18748 |
0.19 |
| chr11_66168455_66168617 | 0.15 |
Dnah9 |
dynein, axonemal, heavy chain 9 |
3 |
0.91 |
| chr3_21983890_21984041 | 0.15 |
Gm43674 |
predicted gene 43674 |
14503 |
0.21 |
| chr5_92097101_92097294 | 0.15 |
G3bp2 |
GTPase activating protein (SH3 domain) binding protein 2 |
13478 |
0.12 |
| chr14_16496682_16496846 | 0.15 |
Top2b |
topoisomerase (DNA) II beta |
71336 |
0.1 |
| chrX_12042851_12043040 | 0.15 |
Bcor |
BCL6 interacting corepressor |
37608 |
0.18 |
| chr17_34238737_34238896 | 0.15 |
H2-Ob |
histocompatibility 2, O region beta locus |
87 |
0.92 |
| chr1_92113054_92113272 | 0.15 |
Hdac4 |
histone deacetylase 4 |
168 |
0.97 |
| chr19_29046369_29046553 | 0.15 |
1700018L02Rik |
RIKEN cDNA 1700018L02 gene |
216 |
0.87 |
| chr8_93190868_93191185 | 0.15 |
Gm45909 |
predicted gene 45909 |
332 |
0.84 |
| chr8_70469859_70470010 | 0.15 |
Klhl26 |
kelch-like 26 |
5964 |
0.09 |
| chr6_38435070_38435344 | 0.15 |
Ubn2 |
ubinuclein 2 |
1034 |
0.47 |
| chr16_5134848_5135218 | 0.15 |
Ppl |
periplakin |
2612 |
0.19 |
| chr4_122997354_122997619 | 0.14 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
1387 |
0.34 |
| chr13_44328539_44328696 | 0.14 |
Gm29676 |
predicted gene, 29676 |
48168 |
0.12 |
| chr4_126259234_126259772 | 0.14 |
Trappc3 |
trafficking protein particle complex 3 |
2822 |
0.16 |
| chr7_26838778_26838929 | 0.14 |
Cyp2a5 |
cytochrome P450, family 2, subfamily a, polypeptide 5 |
1615 |
0.34 |
| chr11_16911280_16911439 | 0.14 |
Egfr |
epidermal growth factor receptor |
6174 |
0.2 |
| chr16_97511614_97511792 | 0.14 |
Fam3b |
family with sequence similarity 3, member B |
827 |
0.52 |
| chr12_86512537_86512711 | 0.14 |
Esrrb |
estrogen related receptor, beta |
8215 |
0.26 |
| chr4_82469279_82469447 | 0.14 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
29953 |
0.16 |
| chr9_69006751_69006913 | 0.14 |
Rora |
RAR-related orphan receptor alpha |
188709 |
0.03 |
| chr2_132597967_132598125 | 0.13 |
AU019990 |
expressed sequence AU019990 |
149 |
0.95 |
| chr6_38637205_38637362 | 0.13 |
Klrg2 |
killer cell lectin-like receptor subfamily G, member 2 |
41 |
0.98 |
| chr7_126212979_126213138 | 0.13 |
Xpo6 |
exportin 6 |
12557 |
0.12 |
| chr17_34296185_34296361 | 0.13 |
H2-Aa |
histocompatibility 2, class II antigen A, alpha |
8450 |
0.08 |
| chr15_59027576_59027739 | 0.13 |
Mtss1 |
MTSS I-BAR domain containing 1 |
12939 |
0.2 |
| chr14_63411641_63411792 | 0.13 |
Blk |
B lymphoid kinase |
5321 |
0.16 |
| chr1_67134415_67135075 | 0.13 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
11719 |
0.24 |
| chr9_99167096_99167251 | 0.13 |
Pik3cb |
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
26552 |
0.13 |
| chr4_117994950_117995268 | 0.13 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
20073 |
0.14 |
| chr7_40718144_40718310 | 0.13 |
4933402C06Rik |
RIKEN cDNA 4933402C06 gene |
43490 |
0.14 |
| chr10_127571278_127571595 | 0.13 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
10449 |
0.1 |
| chr10_89769233_89769434 | 0.13 |
Uhrf1bp1l |
UHRF1 (ICBP90) binding protein 1-like |
17867 |
0.18 |
| chr10_55257530_55257718 | 0.13 |
Gm46190 |
predicted gene, 46190 |
133262 |
0.06 |
| chr14_25793517_25793703 | 0.13 |
Gm27185 |
predicted gene 27185 |
16271 |
0.13 |
| chr16_97475742_97475907 | 0.13 |
Fam3b |
family with sequence similarity 3, member B |
483 |
0.72 |
| chr11_60220800_60221010 | 0.12 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
301 |
0.83 |
| chr10_93899366_93899640 | 0.12 |
4930471D02Rik |
RIKEN cDNA 4930471D02 gene |
2345 |
0.18 |
| chr6_90553748_90553958 | 0.12 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
3005 |
0.19 |
| chr15_3457404_3457613 | 0.12 |
Ghr |
growth hormone receptor |
14136 |
0.27 |
| chr11_117083257_117083524 | 0.12 |
Gm24323 |
predicted gene, 24323 |
2446 |
0.15 |
| chr15_3725358_3725509 | 0.12 |
Gm4823 |
predicted gene 4823 |
21442 |
0.25 |
| chr11_16814706_16815122 | 0.12 |
Egfros |
epidermal growth factor receptor, opposite strand |
15788 |
0.21 |
| chr16_17489651_17490107 | 0.12 |
Aifm3 |
apoptosis-inducing factor, mitochondrion-associated 3 |
32 |
0.96 |
| chr7_75686656_75686824 | 0.12 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
4206 |
0.2 |
| chr10_53993922_53994073 | 0.12 |
Man1a |
mannosidase 1, alpha |
16882 |
0.23 |
| chr2_147971964_147972189 | 0.12 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
4109 |
0.27 |
| chr11_119607460_119607644 | 0.12 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
3854 |
0.21 |
| chr7_97411671_97412103 | 0.12 |
Thrsp |
thyroid hormone responsive |
5632 |
0.13 |
| chr17_12318423_12318969 | 0.12 |
Map3k4 |
mitogen-activated protein kinase kinase kinase 4 |
52 |
0.63 |
| chr2_129865084_129865235 | 0.12 |
Stk35 |
serine/threonine kinase 35 |
64642 |
0.1 |
| chr9_96726906_96727072 | 0.12 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
2047 |
0.28 |
| chr17_45564691_45565065 | 0.12 |
Slc35b2 |
solute carrier family 35, member B2 |
139 |
0.91 |
| chr10_89460914_89461087 | 0.12 |
Gas2l3 |
growth arrest-specific 2 like 3 |
17033 |
0.22 |
| chr6_49977860_49978030 | 0.12 |
Gm3455 |
predicted gene 3455 |
73241 |
0.11 |
| chr10_104423272_104423453 | 0.12 |
Gm22945 |
predicted gene, 22945 |
47845 |
0.17 |
| chr1_191721016_191721195 | 0.12 |
Lpgat1 |
lysophosphatidylglycerol acyltransferase 1 |
2584 |
0.25 |
| chr17_26633881_26634242 | 0.12 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
1962 |
0.24 |
| chr6_24599661_24599829 | 0.12 |
Lmod2 |
leiomodin 2 (cardiac) |
1983 |
0.26 |
| chr18_60804947_60805111 | 0.12 |
Cd74 |
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
1124 |
0.38 |
| chr9_118101918_118102083 | 0.11 |
Azi2 |
5-azacytidine induced gene 2 |
42944 |
0.13 |
| chr1_31034685_31034850 | 0.11 |
Gm28644 |
predicted gene 28644 |
7152 |
0.17 |
| chr18_12825759_12825954 | 0.11 |
Osbpl1a |
oxysterol binding protein-like 1A |
5949 |
0.2 |
| chr12_72955142_72955307 | 0.11 |
Gm38170 |
predicted gene, 38170 |
4329 |
0.17 |
| chr16_93819131_93819299 | 0.11 |
Dop1b |
DOP1 leucine zipper like protein B |
9781 |
0.13 |
| chr2_79986893_79987051 | 0.11 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
52057 |
0.17 |
| chr16_43328614_43329118 | 0.11 |
Gm15711 |
predicted gene 15711 |
16272 |
0.16 |
| chr3_10041035_10041365 | 0.11 |
Gm38335 |
predicted gene, 38335 |
15661 |
0.15 |
| chr17_46129789_46130129 | 0.11 |
Mrps18a |
mitochondrial ribosomal protein S18A |
4501 |
0.12 |
| chr7_67371889_67372064 | 0.11 |
Mef2a |
myocyte enhancer factor 2A |
851 |
0.44 |
| chr11_119222151_119222324 | 0.11 |
Tbc1d16 |
TBC1 domain family, member 16 |
261 |
0.85 |
| chr6_145866005_145866307 | 0.11 |
Bhlhe41 |
basic helix-loop-helix family, member e41 |
598 |
0.7 |
| chr9_46015412_46015589 | 0.11 |
Sik3 |
SIK family kinase 3 |
2500 |
0.2 |
| chr3_119589038_119589201 | 0.11 |
Gm23432 |
predicted gene, 23432 |
49755 |
0.16 |
| chr1_82189665_82189841 | 0.11 |
Gm9747 |
predicted gene 9747 |
43359 |
0.14 |
| chr1_74807385_74807536 | 0.11 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
14097 |
0.11 |
| chr5_66011974_66012125 | 0.11 |
9130230L23Rik |
RIKEN cDNA 9130230L23 gene |
7764 |
0.12 |
| chr17_84984575_84984726 | 0.11 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
9023 |
0.18 |
| chr7_126280513_126280705 | 0.11 |
Sbk1 |
SH3-binding kinase 1 |
7209 |
0.12 |
| chr2_84639968_84640170 | 0.11 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
1964 |
0.19 |
| chr18_65154932_65155123 | 0.10 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
707 |
0.74 |
| chr15_38569128_38569424 | 0.10 |
Gm29697 |
predicted gene, 29697 |
4720 |
0.14 |
| chr13_46164802_46165009 | 0.10 |
Gm10113 |
predicted gene 10113 |
26141 |
0.22 |
| chr4_108090689_108090857 | 0.10 |
Mir6397 |
microRNA 6397 |
3416 |
0.18 |
| chr4_102517129_102517286 | 0.10 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
46810 |
0.19 |
| chr10_68150861_68151038 | 0.10 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
14323 |
0.25 |
| chr19_60757781_60757932 | 0.10 |
Nanos1 |
nanos C2HC-type zinc finger 1 |
1869 |
0.23 |
| chr14_70629530_70629690 | 0.10 |
Dmtn |
dematin actin binding protein |
544 |
0.65 |
| chr4_129229573_129229757 | 0.10 |
C77080 |
expressed sequence C77080 |
1754 |
0.25 |
| chr19_16579816_16579981 | 0.10 |
Gm50363 |
predicted gene, 50363 |
7430 |
0.23 |
| chr2_26461392_26461595 | 0.10 |
Notch1 |
notch 1 |
1254 |
0.26 |
| chr6_51698327_51698491 | 0.10 |
Gm38811 |
predicted gene, 38811 |
12672 |
0.24 |
| chr8_95850398_95850569 | 0.10 |
Slc38a7 |
solute carrier family 38, member 7 |
3039 |
0.13 |
| chr12_16586552_16586706 | 0.10 |
Lpin1 |
lipin 1 |
3091 |
0.3 |
| chr7_34265793_34265944 | 0.10 |
4931406P16Rik |
RIKEN cDNA 4931406P16 gene |
8649 |
0.1 |
| chr3_97225695_97225846 | 0.10 |
Bcl9 |
B cell CLL/lymphoma 9 |
1561 |
0.36 |
| chr2_167300044_167300203 | 0.10 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
5575 |
0.18 |
| chr17_34285962_34286127 | 0.10 |
H2-Aa |
histocompatibility 2, class II antigen A, alpha |
1733 |
0.17 |
| chr6_88898420_88898581 | 0.10 |
Mcm2 |
minichromosome maintenance complex component 2 |
177 |
0.91 |
| chr5_54007502_54007692 | 0.10 |
Stim2 |
stromal interaction molecule 2 |
9032 |
0.25 |
| chr15_29977118_29977282 | 0.10 |
Gm24554 |
predicted gene, 24554 |
26934 |
0.14 |
| chr17_47009170_47009329 | 0.10 |
Ubr2 |
ubiquitin protein ligase E3 component n-recognin 2 |
1230 |
0.43 |
| chr12_77500760_77500939 | 0.10 |
Gm48177 |
predicted gene, 48177 |
25549 |
0.17 |
| chr18_75190731_75190882 | 0.10 |
Gm8807 |
predicted gene 8807 |
7992 |
0.23 |
| chr12_81190858_81191019 | 0.10 |
Mir3067 |
microRNA 3067 |
24787 |
0.17 |
| chr6_17463758_17464495 | 0.10 |
Met |
met proto-oncogene |
19 |
0.98 |
| chr11_81413147_81413311 | 0.10 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
28216 |
0.24 |
| chr4_135326152_135326330 | 0.10 |
Srrm1 |
serine/arginine repetitive matrix 1 |
982 |
0.41 |
| chr1_31051365_31051516 | 0.10 |
Gm28644 |
predicted gene 28644 |
9521 |
0.17 |
| chr17_45642163_45642318 | 0.09 |
Capn11 |
calpain 11 |
17057 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0014854 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0052760 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |