Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
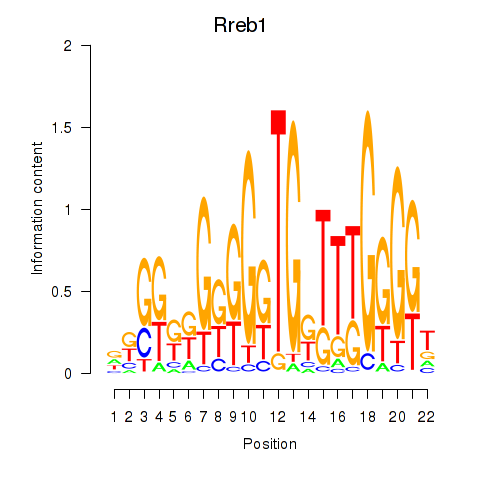
Results for Rreb1
Z-value: 2.10
Transcription factors associated with Rreb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rreb1
|
ENSMUSG00000039087.10 | ras responsive element binding protein 1 |
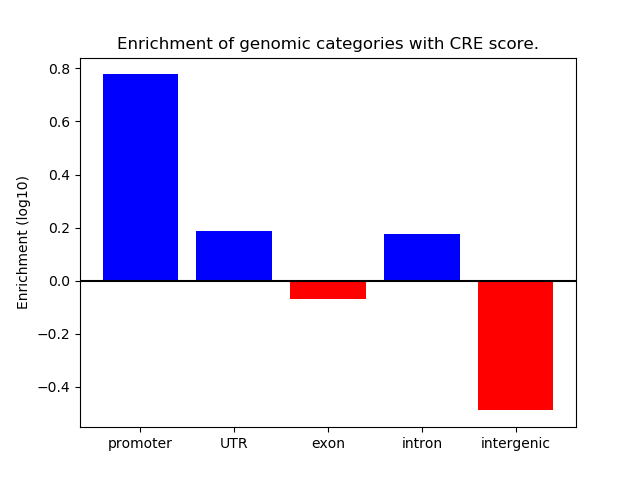
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_37779207_37779426 | Rreb1 | 916 | 0.514807 | 0.92 | 1.0e-02 | Click! |
| chr13_37912095_37912275 | Rreb1 | 23314 | 0.185962 | -0.88 | 2.2e-02 | Click! |
| chr13_37845722_37845913 | Rreb1 | 12117 | 0.192690 | -0.86 | 3.0e-02 | Click! |
| chr13_37866047_37866235 | Rreb1 | 8207 | 0.215890 | 0.83 | 4.0e-02 | Click! |
| chr13_37827559_37827750 | Rreb1 | 261 | 0.917719 | 0.83 | 4.3e-02 | Click! |
Activity of the Rreb1 motif across conditions
Conditions sorted by the z-value of the Rreb1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
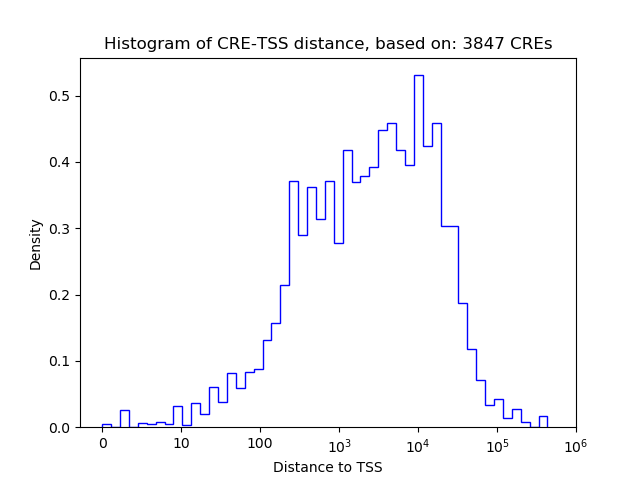
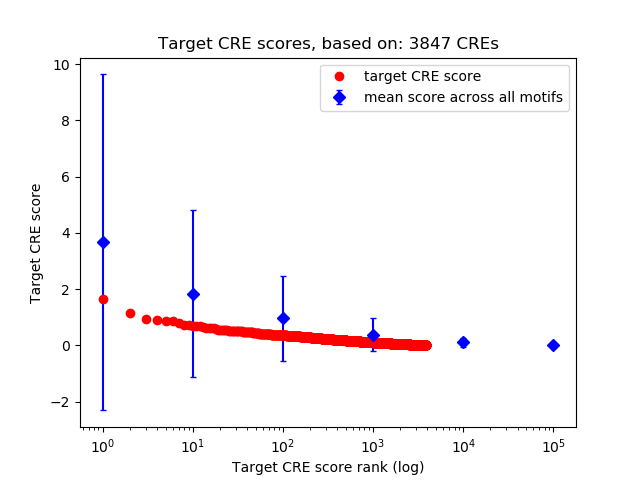
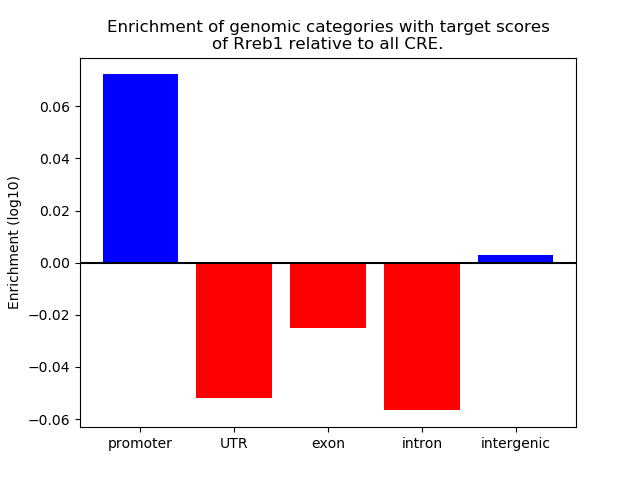
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_93630535_93630727 | 1.64 |
Gm15622 |
predicted gene 15622 |
5249 |
0.17 |
| chr1_39192284_39192607 | 1.16 |
Npas2 |
neuronal PAS domain protein 2 |
1286 |
0.45 |
| chr17_31872001_31872152 | 0.93 |
Sik1 |
salt inducible kinase 1 |
16272 |
0.13 |
| chr10_59786684_59786837 | 0.89 |
Gm17059 |
predicted gene 17059 |
13494 |
0.14 |
| chr1_72827270_72827421 | 0.88 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
2023 |
0.37 |
| chr19_46133855_46134398 | 0.88 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
2229 |
0.19 |
| chr4_141276769_141276920 | 0.78 |
Gm13056 |
predicted gene 13056 |
1595 |
0.24 |
| chr8_126707171_126707322 | 0.74 |
Gm45805 |
predicted gene 45805 |
51088 |
0.15 |
| chr13_60479839_60480430 | 0.72 |
Gm48500 |
predicted gene, 48500 |
1164 |
0.45 |
| chr10_85186626_85186777 | 0.69 |
Cry1 |
cryptochrome 1 (photolyase-like) |
1637 |
0.39 |
| chr5_150227882_150228033 | 0.68 |
Gm36378 |
predicted gene, 36378 |
15906 |
0.19 |
| chr3_85859423_85859574 | 0.68 |
Gm37240 |
predicted gene, 37240 |
27870 |
0.1 |
| chr9_121906088_121906239 | 0.65 |
Ackr2 |
atypical chemokine receptor 2 |
1 |
0.95 |
| chrX_87238307_87238458 | 0.62 |
Gm8855 |
predicted gene 8855 |
435487 |
0.01 |
| chr2_31492850_31493284 | 0.62 |
Ass1 |
argininosuccinate synthetase 1 |
4705 |
0.21 |
| chr6_91707022_91707254 | 0.62 |
Gm45828 |
predicted gene 45828 |
2268 |
0.2 |
| chr11_33931902_33932233 | 0.61 |
Kcnmb1 |
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
30946 |
0.19 |
| chr17_84896257_84896408 | 0.57 |
Gm49982 |
predicted gene, 49982 |
6237 |
0.19 |
| chr4_62780734_62780897 | 0.56 |
Gm24117 |
predicted gene, 24117 |
31613 |
0.15 |
| chr16_37867529_37867680 | 0.56 |
Lrrc58 |
leucine rich repeat containing 58 |
785 |
0.54 |
| chr12_21141056_21141213 | 0.55 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
29180 |
0.17 |
| chr4_148712874_148713025 | 0.54 |
Gm13203 |
predicted gene 13203 |
754 |
0.66 |
| chr4_137874478_137874749 | 0.53 |
Gm13012 |
predicted gene 13012 |
8799 |
0.2 |
| chr6_125486009_125486160 | 0.53 |
Gm28809 |
predicted gene 28809 |
4140 |
0.17 |
| chr8_3393831_3393982 | 0.52 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
833 |
0.6 |
| chr1_120118168_120118368 | 0.52 |
Dbi |
diazepam binding inhibitor |
1870 |
0.3 |
| chr12_108837829_108837980 | 0.52 |
Slc25a47 |
solute carrier family 25, member 47 |
840 |
0.31 |
| chr12_59194921_59195102 | 0.52 |
Fbxo33 |
F-box protein 33 |
9425 |
0.15 |
| chr7_31053117_31053317 | 0.51 |
Fxyd1 |
FXYD domain-containing ion transport regulator 1 |
95 |
0.91 |
| chr2_60863337_60863488 | 0.51 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
18026 |
0.26 |
| chr3_97655610_97655761 | 0.51 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
2508 |
0.2 |
| chr6_23392505_23392811 | 0.50 |
Cadps2 |
Ca2+-dependent activator protein for secretion 2 |
36739 |
0.19 |
| chr2_157571407_157571591 | 0.50 |
Blcap |
bladder cancer associated protein |
225 |
0.89 |
| chr15_88820456_88820732 | 0.50 |
Creld2 |
cysteine-rich with EGF-like domains 2 |
948 |
0.39 |
| chr8_123812047_123812213 | 0.49 |
Rab4a |
RAB4A, member RAS oncogene family |
6062 |
0.1 |
| chr19_30171407_30171581 | 0.49 |
Gldc |
glycine decarboxylase |
3935 |
0.23 |
| chr17_34955918_34956069 | 0.49 |
Gm8696 |
predicted gene 8696 |
1281 |
0.14 |
| chr15_85716911_85717062 | 0.49 |
Mirlet7b |
microRNA let7b |
9667 |
0.13 |
| chr9_121912346_121912498 | 0.48 |
Cyp8b1 |
cytochrome P450, family 8, subfamily b, polypeptide 1 |
3883 |
0.1 |
| chr11_101668758_101669043 | 0.47 |
Arl4d |
ADP-ribosylation factor-like 4D |
3359 |
0.12 |
| chr13_45963217_45963398 | 0.47 |
Atxn1 |
ataxin 1 |
1650 |
0.33 |
| chr1_191946290_191946441 | 0.46 |
Rd3 |
retinal degeneration 3 |
31005 |
0.1 |
| chr15_103364813_103364984 | 0.46 |
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
1850 |
0.21 |
| chr13_60607775_60607969 | 0.46 |
Dapk1 |
death associated protein kinase 1 |
5640 |
0.18 |
| chr7_63896677_63896828 | 0.46 |
Gm27252 |
predicted gene 27252 |
219 |
0.91 |
| chr2_103850440_103850628 | 0.46 |
Gm13879 |
predicted gene 13879 |
6878 |
0.09 |
| chr14_73630476_73630939 | 0.45 |
Gm21750 |
predicted gene, 21750 |
30518 |
0.15 |
| chr10_43742623_43743061 | 0.44 |
Gm40634 |
predicted gene, 40634 |
245 |
0.89 |
| chr9_63550824_63550989 | 0.44 |
Gm16759 |
predicted gene, 16759 |
19554 |
0.18 |
| chr3_89271513_89271664 | 0.42 |
Slc50a1 |
solute carrier family 50 (sugar transporter), member 1 |
1018 |
0.25 |
| chr7_112300238_112300389 | 0.42 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
15716 |
0.27 |
| chr3_84258598_84259173 | 0.42 |
Trim2 |
tripartite motif-containing 2 |
484 |
0.86 |
| chr2_60383459_60383659 | 0.42 |
Ly75 |
lymphocyte antigen 75 |
256 |
0.91 |
| chr10_94159643_94159802 | 0.42 |
Nr2c1 |
nuclear receptor subfamily 2, group C, member 1 |
3436 |
0.18 |
| chr10_95780270_95780625 | 0.42 |
4732465J04Rik |
RIKEN cDNA 4732465J04 gene |
1530 |
0.29 |
| chr16_38246497_38246731 | 0.42 |
Gsk3b |
glycogen synthase kinase 3 beta |
38576 |
0.11 |
| chr3_51255928_51256079 | 0.42 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr4_135326152_135326330 | 0.41 |
Srrm1 |
serine/arginine repetitive matrix 1 |
982 |
0.41 |
| chr4_144908630_144908793 | 0.41 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
10168 |
0.21 |
| chr2_163750130_163750300 | 0.41 |
Ada |
adenosine deaminase |
24 |
0.98 |
| chr10_99222701_99222877 | 0.41 |
Gm34574 |
predicted gene, 34574 |
277 |
0.85 |
| chr3_52258108_52258303 | 0.40 |
Foxo1 |
forkhead box O1 |
10131 |
0.12 |
| chr11_102142277_102142428 | 0.40 |
Nags |
N-acetylglutamate synthase |
3161 |
0.11 |
| chr15_7070249_7070660 | 0.40 |
Lifr |
LIF receptor alpha |
20160 |
0.25 |
| chr14_30891050_30891367 | 0.40 |
Itih4 |
inter alpha-trypsin inhibitor, heavy chain 4 |
4687 |
0.13 |
| chr19_46714530_46714681 | 0.40 |
As3mt |
arsenic (+3 oxidation state) methyltransferase |
2633 |
0.2 |
| chr11_4115460_4115726 | 0.40 |
Sec14l2 |
SEC14-like lipid binding 2 |
3103 |
0.12 |
| chr17_12682270_12682463 | 0.39 |
Slc22a1 |
solute carrier family 22 (organic cation transporter), member 1 |
6537 |
0.17 |
| chr12_76554293_76554484 | 0.39 |
AI463170 |
expressed sequence AI463170 |
6445 |
0.14 |
| chr1_131275281_131275570 | 0.39 |
Ikbke |
inhibitor of kappaB kinase epsilon |
1288 |
0.32 |
| chr5_125313647_125313811 | 0.39 |
Scarb1 |
scavenger receptor class B, member 1 |
2903 |
0.2 |
| chr18_54969498_54969696 | 0.39 |
Zfp608 |
zinc finger protein 608 |
12840 |
0.2 |
| chr8_44972192_44972436 | 0.39 |
Fat1 |
FAT atypical cadherin 1 |
22100 |
0.17 |
| chr5_122520868_122521019 | 0.38 |
Gm22965 |
predicted gene, 22965 |
14154 |
0.09 |
| chr10_42764114_42764349 | 0.38 |
Sec63 |
SEC63-like (S. cerevisiae) |
2425 |
0.25 |
| chr2_31505484_31505674 | 0.38 |
Ass1 |
argininosuccinate synthetase 1 |
4853 |
0.2 |
| chr12_76289294_76289480 | 0.38 |
Mthfd1 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
3678 |
0.14 |
| chr17_34398487_34398638 | 0.38 |
BC051142 |
cDNA sequence BC051142 |
258 |
0.59 |
| chr9_78192592_78192962 | 0.38 |
Gsta4 |
glutathione S-transferase, alpha 4 |
830 |
0.46 |
| chr8_123757273_123757470 | 0.38 |
4732419C18Rik |
RIKEN cDNA 4732419C18 gene |
2645 |
0.11 |
| chr4_133184820_133185010 | 0.37 |
Wasf2 |
WAS protein family, member 2 |
4103 |
0.15 |
| chr6_122595445_122595623 | 0.37 |
Apobec1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
4593 |
0.12 |
| chr13_93633844_93634026 | 0.37 |
Bhmt |
betaine-homocysteine methyltransferase |
3631 |
0.19 |
| chr12_54967535_54967800 | 0.37 |
Gm27014 |
predicted gene, 27014 |
4603 |
0.15 |
| chr2_103867069_103867263 | 0.37 |
Gm13876 |
predicted gene 13876 |
21158 |
0.08 |
| chr3_66035020_66035198 | 0.37 |
Gm6546 |
predicted gene 6546 |
3590 |
0.17 |
| chr11_109651940_109652143 | 0.37 |
Prkar1a |
protein kinase, cAMP dependent regulatory, type I, alpha |
1120 |
0.46 |
| chr17_80325457_80325860 | 0.37 |
Arhgef33 |
Rho guanine nucleotide exchange factor (GEF) 33 |
18233 |
0.15 |
| chr9_30897343_30897518 | 0.37 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
12227 |
0.22 |
| chr12_8507138_8507301 | 0.36 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
3982 |
0.18 |
| chr10_84907603_84907783 | 0.36 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
9923 |
0.24 |
| chr18_38856344_38856590 | 0.36 |
Fgf1 |
fibroblast growth factor 1 |
2281 |
0.33 |
| chr7_28595870_28596035 | 0.36 |
Pak4 |
p21 (RAC1) activated kinase 4 |
2134 |
0.15 |
| chr10_18395038_18395462 | 0.36 |
Nhsl1 |
NHS-like 1 |
4928 |
0.26 |
| chr19_3895426_3895581 | 0.36 |
Tcirg1 |
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
1727 |
0.17 |
| chr2_170405048_170405199 | 0.36 |
Bcas1os1 |
breast carcinoma amplified sequence 1, opposite strand 1 |
58 |
0.96 |
| chr5_124425906_124426318 | 0.35 |
Sbno1 |
strawberry notch 1 |
111 |
0.73 |
| chr5_150227352_150227751 | 0.35 |
Gm36378 |
predicted gene, 36378 |
16312 |
0.19 |
| chr5_120495968_120496124 | 0.35 |
Plbd2 |
phospholipase B domain containing 2 |
7555 |
0.11 |
| chr6_72624193_72624362 | 0.35 |
Gm15401 |
predicted gene 15401 |
5011 |
0.09 |
| chr14_63942095_63942273 | 0.35 |
Sox7 |
SRY (sex determining region Y)-box 7 |
1489 |
0.38 |
| chr3_121446410_121446561 | 0.35 |
Cnn3 |
calponin 3, acidic |
3281 |
0.18 |
| chr16_21818058_21818209 | 0.35 |
Map3k13 |
mitogen-activated protein kinase kinase kinase 13 |
7809 |
0.12 |
| chr9_14174315_14174466 | 0.35 |
Gm47564 |
predicted gene, 47564 |
29693 |
0.15 |
| chr1_156073065_156073216 | 0.35 |
Tor1aip2 |
torsin A interacting protein 2 |
9927 |
0.17 |
| chr14_29998857_29999427 | 0.35 |
Il17rb |
interleukin 17 receptor B |
683 |
0.54 |
| chr11_43643345_43643496 | 0.35 |
Gm12149 |
predicted gene 12149 |
27091 |
0.12 |
| chr6_145343654_145343918 | 0.35 |
Gm23498 |
predicted gene, 23498 |
23072 |
0.11 |
| chr17_34722831_34722982 | 0.35 |
Cyp21a2-ps |
cytochrome P450, family 21, subfamily a, polypeptide 2 pseudogene |
654 |
0.44 |
| chr18_20990195_20990346 | 0.35 |
Rnf138 |
ring finger protein 138 |
11071 |
0.2 |
| chrX_13275719_13275870 | 0.35 |
Ddx3x |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked |
5176 |
0.14 |
| chr5_125323526_125323720 | 0.35 |
Gm42633 |
predicted gene 42633 |
2179 |
0.22 |
| chr8_46479835_46479986 | 0.35 |
Gm45244 |
predicted gene 45244 |
6320 |
0.16 |
| chr4_136138924_136139083 | 0.35 |
Gm26001 |
predicted gene, 26001 |
1507 |
0.3 |
| chr11_75279612_75279763 | 0.35 |
Gm47300 |
predicted gene, 47300 |
38921 |
0.1 |
| chr1_72810299_72810480 | 0.34 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
14114 |
0.21 |
| chr1_105905887_105906083 | 0.34 |
Gm37779 |
predicted gene, 37779 |
14973 |
0.17 |
| chr11_120009704_120009855 | 0.34 |
Aatk |
apoptosis-associated tyrosine kinase |
2629 |
0.15 |
| chr4_123093071_123093222 | 0.34 |
Gm12923 |
predicted gene 12923 |
882 |
0.46 |
| chr17_47593664_47594464 | 0.34 |
Ccnd3 |
cyclin D3 |
249 |
0.86 |
| chr9_57638948_57639112 | 0.34 |
Csk |
c-src tyrosine kinase |
4963 |
0.12 |
| chr2_127071034_127071236 | 0.34 |
Blvra |
biliverdin reductase A |
442 |
0.79 |
| chr5_8056810_8056986 | 0.34 |
Sri |
sorcin |
290 |
0.85 |
| chr15_59775397_59775598 | 0.34 |
Gm19510 |
predicted gene, 19510 |
19462 |
0.24 |
| chr8_12800203_12800354 | 0.34 |
Atp11a |
ATPase, class VI, type 11A |
11138 |
0.16 |
| chr8_46750315_46750466 | 0.33 |
Irf2 |
interferon regulatory factor 2 |
10124 |
0.14 |
| chr13_113022309_113022460 | 0.33 |
Cdc20b |
cell division cycle 20B |
12727 |
0.08 |
| chr12_110867720_110867879 | 0.33 |
Mpc1-ps |
mitochondrial pyruvate carrier 1, pseudogene |
8905 |
0.09 |
| chr14_40930703_40931117 | 0.33 |
Tspan14 |
tetraspanin 14 |
3406 |
0.26 |
| chr6_99175134_99175460 | 0.33 |
Foxp1 |
forkhead box P1 |
12279 |
0.28 |
| chr1_67039034_67039385 | 0.33 |
Lancl1 |
LanC (bacterial lantibiotic synthetase component C)-like 1 |
337 |
0.9 |
| chr5_141381660_141381822 | 0.33 |
Gm16035 |
predicted gene 16035 |
815 |
0.63 |
| chr11_116165413_116165564 | 0.33 |
Fbf1 |
Fas (TNFRSF6) binding factor 1 |
395 |
0.67 |
| chr11_46578775_46579093 | 0.33 |
BC053393 |
cDNA sequence BC053393 |
7398 |
0.13 |
| chrX_36644878_36645044 | 0.33 |
Akap17b |
A kinase (PRKA) anchor protein 17B |
135 |
0.96 |
| chr7_35802959_35803128 | 0.33 |
Zfp507 |
zinc finger protein 507 |
40 |
0.88 |
| chr2_30418585_30418853 | 0.33 |
Ptpa |
protein phosphatase 2 protein activator |
2417 |
0.15 |
| chr12_8433741_8433892 | 0.33 |
Gm48076 |
predicted gene, 48076 |
21102 |
0.15 |
| chr11_120521031_120521212 | 0.32 |
Gm11789 |
predicted gene 11789 |
9433 |
0.06 |
| chr4_107020194_107020543 | 0.32 |
Gm12786 |
predicted gene 12786 |
2131 |
0.26 |
| chr18_65223520_65223956 | 0.32 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
15854 |
0.15 |
| chr8_57488604_57488764 | 0.32 |
2500002B13Rik |
RIKEN cDNA 2500002B13 gene |
630 |
0.45 |
| chr11_53359050_53359204 | 0.32 |
Aff4 |
AF4/FMR2 family, member 4 |
8294 |
0.11 |
| chr10_81600790_81601143 | 0.32 |
Tle6 |
transducin-like enhancer of split 6 |
66 |
0.93 |
| chr15_88859255_88859406 | 0.32 |
Pim3 |
proviral integration site 3 |
2856 |
0.19 |
| chr14_70154015_70154175 | 0.32 |
Ccar2 |
cell cycle activator and apoptosis regulator 2 |
284 |
0.85 |
| chr18_46336206_46336490 | 0.32 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
4081 |
0.17 |
| chr13_93636362_93636873 | 0.32 |
Bhmt |
betaine-homocysteine methyltransferase |
949 |
0.5 |
| chr7_118739850_118740027 | 0.32 |
Vps35l |
VPS35 endosomal protein sorting factor like |
288 |
0.8 |
| chr15_76558868_76559133 | 0.32 |
Adck5 |
aarF domain containing kinase 5 |
17358 |
0.06 |
| chr7_3230802_3230999 | 0.32 |
Mir295 |
microRNA 295 |
10127 |
0.06 |
| chr1_180161827_180161978 | 0.32 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
895 |
0.53 |
| chr6_117167479_117167672 | 0.31 |
Cxcl12 |
chemokine (C-X-C motif) ligand 12 |
960 |
0.54 |
| chr1_39949768_39950072 | 0.31 |
Map4k4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
14235 |
0.22 |
| chr17_34605267_34605661 | 0.31 |
Agpat1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
397 |
0.55 |
| chr6_48595035_48595341 | 0.31 |
Repin1 |
replication initiator 1 |
869 |
0.27 |
| chr18_20666842_20666993 | 0.31 |
Ttr |
transthyretin |
1637 |
0.28 |
| chr11_79224332_79224483 | 0.31 |
Gm11200 |
predicted gene 11200 |
11611 |
0.17 |
| chr6_125309060_125309481 | 0.31 |
Ltbr |
lymphotoxin B receptor |
295 |
0.81 |
| chr1_67140040_67140230 | 0.31 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
17109 |
0.23 |
| chr17_34836259_34836412 | 0.31 |
Stk19 |
serine/threonine kinase 19 |
130 |
0.8 |
| chrX_7656942_7657153 | 0.30 |
Gm14703 |
predicted gene 14703 |
211 |
0.46 |
| chr11_5486133_5486285 | 0.30 |
Gm11962 |
predicted gene 11962 |
9237 |
0.14 |
| chr19_46976168_46976319 | 0.30 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
6675 |
0.17 |
| chr9_62361596_62361747 | 0.30 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
5267 |
0.23 |
| chr18_46603768_46603972 | 0.30 |
Eif1a |
eukaryotic translation initiation factor 1A |
5856 |
0.15 |
| chr3_156999583_156999772 | 0.30 |
Gm43527 |
predicted gene 43527 |
740 |
0.74 |
| chr19_37702993_37703212 | 0.30 |
Cyp26a1 |
cytochrome P450, family 26, subfamily a, polypeptide 1 |
5017 |
0.2 |
| chr7_98413497_98413672 | 0.30 |
Gm44507 |
predicted gene 44507 |
2040 |
0.24 |
| chr9_48724021_48724519 | 0.30 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
111675 |
0.06 |
| chr12_111819707_111819858 | 0.30 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
3526 |
0.14 |
| chr2_31490719_31491007 | 0.30 |
Ass1 |
argininosuccinate synthetase 1 |
6909 |
0.19 |
| chr16_10541317_10541611 | 0.29 |
Dexi |
dexamethasone-induced transcript |
1590 |
0.35 |
| chr6_51362990_51363148 | 0.29 |
Gm6559 |
predicted gene 6559 |
16641 |
0.18 |
| chr4_147847988_147848325 | 0.29 |
Zfp933 |
zinc finger protein 933 |
210 |
0.88 |
| chr17_31852879_31853048 | 0.29 |
Sik1 |
salt inducible kinase 1 |
1388 |
0.38 |
| chr10_96924578_96924746 | 0.29 |
Gm33981 |
predicted gene, 33981 |
34209 |
0.17 |
| chr5_135029120_135029476 | 0.29 |
Stx1a |
syntaxin 1A (brain) |
5738 |
0.08 |
| chr2_31451697_31451848 | 0.29 |
Hmcn2 |
hemicentin 2 |
2936 |
0.26 |
| chr8_120409781_120409965 | 0.29 |
Gm22715 |
predicted gene, 22715 |
33676 |
0.16 |
| chr3_84813766_84813917 | 0.29 |
Fbxw7 |
F-box and WD-40 domain protein 7 |
1427 |
0.53 |
| chr9_65243868_65244187 | 0.29 |
Cilp |
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
21153 |
0.1 |
| chr4_63557379_63557840 | 0.29 |
Tmem268 |
transmembrane protein 268 |
1172 |
0.35 |
| chr15_80225601_80225752 | 0.29 |
4930588D02Rik |
RIKEN cDNA 4930588D02 gene |
2738 |
0.15 |
| chr1_191956791_191956971 | 0.29 |
Rd3 |
retinal degeneration 3 |
20489 |
0.11 |
| chr1_184871816_184871971 | 0.29 |
C130074G19Rik |
RIKEN cDNA C130074G19 gene |
11325 |
0.16 |
| chr6_72787755_72787940 | 0.29 |
Tcf7l1 |
transcription factor 7 like 1 (T cell specific, HMG box) |
751 |
0.63 |
| chr18_4844934_4845219 | 0.29 |
Gm10556 |
predicted gene 10556 |
32590 |
0.2 |
| chr10_111335589_111335748 | 0.28 |
Gm40761 |
predicted gene, 40761 |
1456 |
0.43 |
| chr8_126660866_126661445 | 0.28 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
67169 |
0.11 |
| chr17_45595822_45596123 | 0.28 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
15 |
0.95 |
| chr14_16248790_16249022 | 0.28 |
Oxsm |
3-oxoacyl-ACP synthase, mitochondrial |
268 |
0.58 |
| chr10_62413042_62413209 | 0.28 |
Hkdc1 |
hexokinase domain containing 1 |
1321 |
0.36 |
| chr7_100011175_100011513 | 0.28 |
Chrdl2 |
chordin-like 2 |
1430 |
0.34 |
| chr1_39025074_39025225 | 0.28 |
Pdcl3 |
phosducin-like 3 |
29989 |
0.13 |
| chr8_44970573_44970724 | 0.28 |
Fat1 |
FAT atypical cadherin 1 |
20434 |
0.18 |
| chr4_123663878_123664029 | 0.28 |
Macf1 |
microtubule-actin crosslinking factor 1 |
799 |
0.5 |
| chr12_111373260_111373430 | 0.28 |
Cdc42bpb |
CDC42 binding protein kinase beta |
4274 |
0.17 |
| chr17_31637083_31637756 | 0.28 |
Cbs |
cystathionine beta-synthase |
181 |
0.89 |
| chr18_35598169_35598395 | 0.28 |
Gm50163 |
predicted gene, 50163 |
291 |
0.49 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 | 0.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.4 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.1 | 0.5 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.1 | 0.7 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.4 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.3 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.6 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.2 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.2 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.1 | 0.3 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 | 0.3 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.0 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 0.0 | 0.3 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.2 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.3 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0048242 | epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:1902661 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0070587 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.2 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0097090 | presynaptic membrane organization(GO:0097090) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:1903659 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.0 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0090239 | regulation of histone H4 acetylation(GO:0090239) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.0 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.0 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.2 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.2 | GO:0034340 | response to type I interferon(GO:0034340) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.4 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0034824 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.0 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |