Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
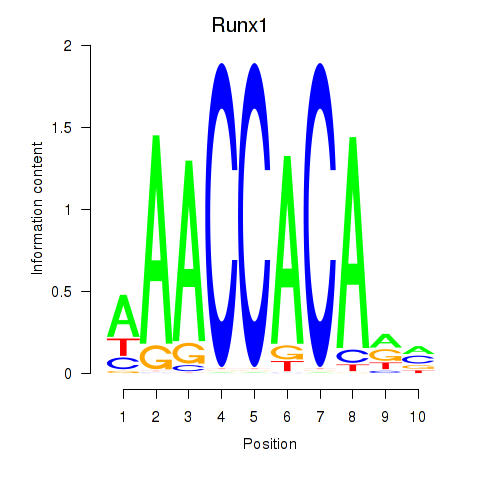
Results for Runx1
Z-value: 0.97
Transcription factors associated with Runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx1
|
ENSMUSG00000022952.10 | runt related transcription factor 1 |
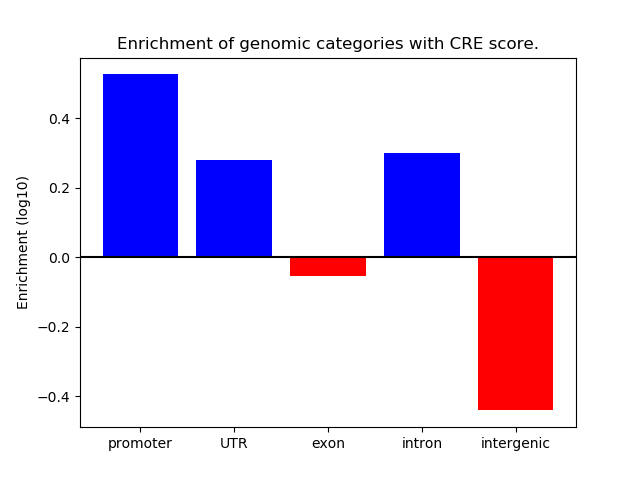
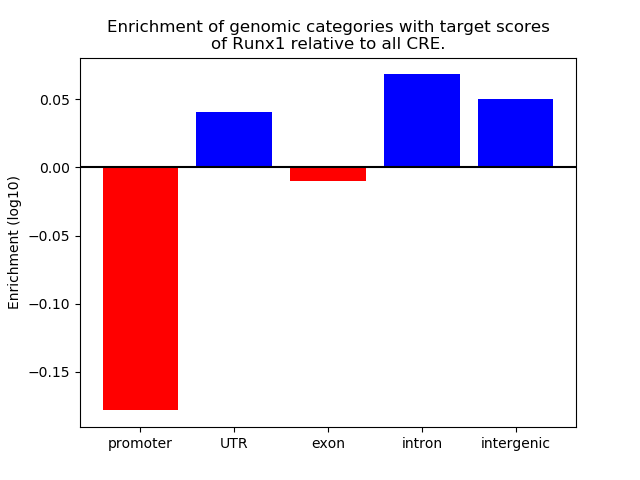
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_92698486_92698672 | Runx1 | 1251 | 0.565237 | -0.74 | 9.2e-02 | Click! |
| chr16_92715564_92715731 | Runx1 | 18319 | 0.239675 | -0.63 | 1.8e-01 | Click! |
| chr16_92697311_92697473 | Runx1 | 64 | 0.982897 | 0.47 | 3.4e-01 | Click! |
| chr16_92696728_92696879 | Runx1 | 522 | 0.845725 | 0.25 | 6.4e-01 | Click! |
| chr16_92826008_92826183 | Runx1 | 21 | 0.984518 | 0.23 | 6.7e-01 | Click! |
Activity of the Runx1 motif across conditions
Conditions sorted by the z-value of the Runx1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
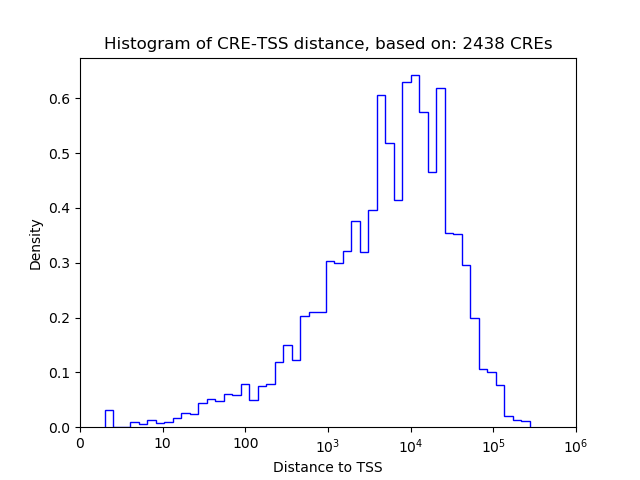
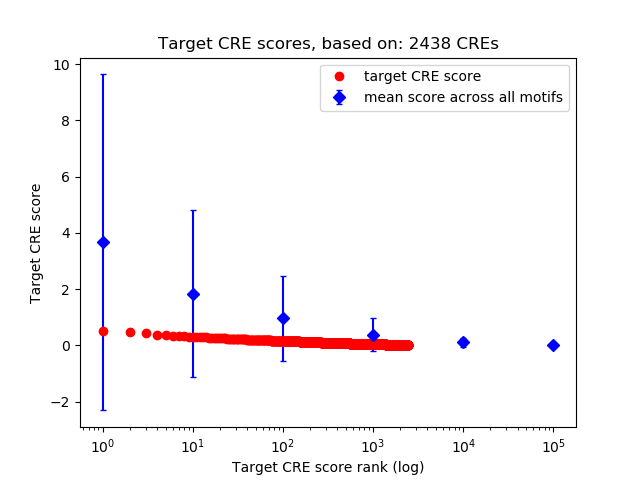
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_27176664_27176830 | 0.51 |
Gm35438 |
predicted gene, 35438 |
15262 |
0.21 |
| chr8_111821313_111821477 | 0.46 |
Cfdp1 |
craniofacial development protein 1 |
32896 |
0.13 |
| chr5_66114850_66115151 | 0.42 |
Rbm47 |
RNA binding motif protein 47 |
16809 |
0.11 |
| chr13_59893097_59893271 | 0.36 |
Gm19866 |
predicted gene, 19866 |
13935 |
0.13 |
| chr13_44293994_44294145 | 0.36 |
Gm29676 |
predicted gene, 29676 |
13620 |
0.2 |
| chr6_55138486_55138637 | 0.35 |
Crhr2 |
corticotropin releasing hormone receptor 2 |
5545 |
0.18 |
| chr5_144126093_144126244 | 0.33 |
Gm26925 |
predicted gene, 26925 |
14396 |
0.14 |
| chr6_127265923_127266082 | 0.31 |
Gm43635 |
predicted gene 43635 |
11358 |
0.12 |
| chr9_94993733_94993884 | 0.30 |
Gm47168 |
predicted gene, 47168 |
1124 |
0.65 |
| chr9_30887209_30887384 | 0.29 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
22361 |
0.2 |
| chr18_39251064_39251253 | 0.29 |
Arhgap26 |
Rho GTPase activating protein 26 |
23790 |
0.21 |
| chr19_8228991_8229142 | 0.29 |
Slc22a29 |
solute carrier family 22. member 29 |
10166 |
0.16 |
| chr5_148936597_148936854 | 0.28 |
Katnal1 |
katanin p60 subunit A-like 1 |
7405 |
0.1 |
| chr7_115686790_115686973 | 0.28 |
Sox6 |
SRY (sex determining region Y)-box 6 |
24516 |
0.27 |
| chr2_132639230_132639433 | 0.28 |
AU019990 |
expressed sequence AU019990 |
6550 |
0.14 |
| chr2_4318191_4318342 | 0.27 |
Gm36988 |
predicted gene, 36988 |
1787 |
0.31 |
| chr19_26748788_26748982 | 0.27 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
353 |
0.9 |
| chr14_66199873_66200054 | 0.26 |
Ptk2b |
PTK2 protein tyrosine kinase 2 beta |
13589 |
0.19 |
| chr6_89213939_89214095 | 0.25 |
Gm839 |
predicted gene 839 |
2220 |
0.22 |
| chr2_68884019_68884170 | 0.25 |
Gm37159 |
predicted gene, 37159 |
7272 |
0.15 |
| chr2_102901417_102901572 | 0.25 |
Cd44 |
CD44 antigen |
1 |
0.98 |
| chr13_36324738_36325087 | 0.25 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
2046 |
0.39 |
| chr4_99028512_99028838 | 0.24 |
Angptl3 |
angiopoietin-like 3 |
2279 |
0.28 |
| chr1_45826928_45827079 | 0.24 |
Wdr75 |
WD repeat domain 75 |
4445 |
0.16 |
| chr15_3533289_3533452 | 0.24 |
Ghr |
growth hormone receptor |
48472 |
0.16 |
| chr1_51881730_51881907 | 0.23 |
Gm28323 |
predicted gene 28323 |
2210 |
0.25 |
| chr19_26754124_26754675 | 0.23 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
799 |
0.69 |
| chr2_14130166_14130332 | 0.23 |
Stam |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
595 |
0.69 |
| chr9_41073606_41073757 | 0.22 |
Ubash3b |
ubiquitin associated and SH3 domain containing, B |
5825 |
0.19 |
| chr1_35889876_35890029 | 0.22 |
1110002O04Rik |
RIKEN cDNA 1110002O04 gene |
10107 |
0.21 |
| chr18_61719777_61719946 | 0.22 |
Grpel2 |
GrpE-like 2, mitochondrial |
639 |
0.59 |
| chr4_41493635_41493797 | 0.22 |
Myorg |
myogenesis regulating glycosidase (putative) |
8952 |
0.1 |
| chr14_51105673_51105825 | 0.22 |
Eddm3b |
epididymal protein 3B |
8677 |
0.08 |
| chr8_115761720_115761892 | 0.22 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
54012 |
0.15 |
| chr11_84476404_84476555 | 0.22 |
Aatf |
apoptosis antagonizing transcription factor |
15071 |
0.21 |
| chr11_38681374_38681545 | 0.21 |
Gm23520 |
predicted gene, 23520 |
116818 |
0.07 |
| chr15_10972894_10973115 | 0.21 |
Amacr |
alpha-methylacyl-CoA racemase |
8752 |
0.15 |
| chr6_88995214_88995375 | 0.21 |
4933427D06Rik |
RIKEN cDNA 4933427D06 gene |
44611 |
0.12 |
| chr1_128016251_128016402 | 0.21 |
Zranb3 |
zinc finger, RAN-binding domain containing 3 |
2367 |
0.22 |
| chr3_108134800_108134966 | 0.21 |
Gnai3 |
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
11263 |
0.09 |
| chr8_86538609_86538876 | 0.21 |
Abcc12 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
3995 |
0.27 |
| chr13_8591659_8591833 | 0.20 |
Gm48262 |
predicted gene, 48262 |
38726 |
0.19 |
| chr2_101797736_101797887 | 0.19 |
Prr5l |
proline rich 5 like |
78 |
0.98 |
| chr1_106231521_106231673 | 0.19 |
Gm38235 |
predicted gene, 38235 |
12283 |
0.21 |
| chr13_85126680_85126842 | 0.19 |
Gm4076 |
predicted gene 4076 |
753 |
0.65 |
| chr6_128646065_128646273 | 0.19 |
Gm5884 |
predicted pseudogene 5884 |
1256 |
0.23 |
| chr19_43949859_43950010 | 0.19 |
Dnmbp |
dynamin binding protein |
9743 |
0.13 |
| chr14_76310421_76310583 | 0.19 |
2900040C04Rik |
RIKEN cDNA 2900040C04 gene |
59045 |
0.12 |
| chr13_100849368_100849536 | 0.19 |
Gm29502 |
predicted gene 29502 |
2019 |
0.24 |
| chr18_12230939_12231163 | 0.19 |
Npc1 |
NPC intracellular cholesterol transporter 1 |
5275 |
0.17 |
| chr3_59114068_59114252 | 0.19 |
Gpr171 |
G protein-coupled receptor 171 |
12339 |
0.16 |
| chr17_31473791_31473953 | 0.19 |
Pde9a |
phosphodiesterase 9A |
2820 |
0.14 |
| chr7_122665413_122665987 | 0.19 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
4792 |
0.21 |
| chr11_106366472_106366631 | 0.19 |
Prr29 |
proline rich 29 |
1079 |
0.4 |
| chr1_138967552_138967703 | 0.19 |
Dennd1b |
DENN/MADD domain containing 1B |
93 |
0.91 |
| chr9_83102256_83102418 | 0.18 |
Gm38398 |
predicted gene, 38398 |
4113 |
0.16 |
| chr5_51969354_51969505 | 0.18 |
Gm43175 |
predicted gene 43175 |
58498 |
0.11 |
| chr4_105104623_105104776 | 0.18 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
5191 |
0.29 |
| chr4_86664611_86664778 | 0.18 |
Plin2 |
perilipin 2 |
1031 |
0.52 |
| chr11_90371567_90371726 | 0.18 |
Hlf |
hepatic leukemia factor |
10289 |
0.26 |
| chr17_13402789_13403038 | 0.18 |
Gm8597 |
predicted gene 8597 |
1278 |
0.29 |
| chr1_128008626_128008793 | 0.18 |
Zranb3 |
zinc finger, RAN-binding domain containing 3 |
9984 |
0.14 |
| chr18_63463560_63463749 | 0.18 |
Gm23581 |
predicted gene, 23581 |
44515 |
0.11 |
| chr2_48766708_48766859 | 0.18 |
Gm13489 |
predicted gene 13489 |
43772 |
0.17 |
| chr13_56354793_56354953 | 0.18 |
Gm10782 |
predicted gene 10782 |
8027 |
0.15 |
| chr19_3293245_3293431 | 0.17 |
Mir6984 |
microRNA 6984 |
4418 |
0.12 |
| chr4_34901497_34901795 | 0.17 |
Cga |
glycoprotein hormones, alpha subunit |
7858 |
0.13 |
| chr7_4080638_4080789 | 0.17 |
Lair1 |
leukocyte-associated Ig-like receptor 1 |
17509 |
0.09 |
| chr17_28696762_28696938 | 0.17 |
Mapk14 |
mitogen-activated protein kinase 14 |
4282 |
0.15 |
| chr3_148423494_148423648 | 0.17 |
Gm43576 |
predicted gene 43576 |
31370 |
0.22 |
| chr1_84241089_84241262 | 0.17 |
Mir6353 |
microRNA 6353 |
22214 |
0.2 |
| chr18_38631612_38631778 | 0.17 |
9630014M24Rik |
RIKEN cDNA 9630014M24 gene |
4408 |
0.19 |
| chr7_125574044_125574195 | 0.17 |
Gm44876 |
predicted gene 44876 |
2002 |
0.27 |
| chr10_80017491_80017661 | 0.17 |
Arhgap45 |
Rho GTPase activating protein 45 |
660 |
0.46 |
| chr2_133427180_133427333 | 0.17 |
A430048G15Rik |
RIKEN cDNA A430048G15 gene |
4839 |
0.26 |
| chr16_4405370_4405807 | 0.17 |
Adcy9 |
adenylate cyclase 9 |
13999 |
0.21 |
| chr4_149282398_149282549 | 0.17 |
Kif1b |
kinesin family member 1B |
25033 |
0.13 |
| chr19_36732071_36732380 | 0.17 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
4428 |
0.24 |
| chr16_31584525_31584676 | 0.17 |
Gm34256 |
predicted gene, 34256 |
8620 |
0.17 |
| chr15_93517178_93517343 | 0.17 |
Prickle1 |
prickle planar cell polarity protein 1 |
2263 |
0.39 |
| chr9_102410773_102410941 | 0.16 |
Gm22894 |
predicted gene, 22894 |
37739 |
0.13 |
| chr3_115789167_115789324 | 0.16 |
Gm9889 |
predicted gene 9889 |
74095 |
0.08 |
| chr14_7821426_7821604 | 0.16 |
Flnb |
filamin, beta |
3558 |
0.19 |
| chr14_66660586_66660748 | 0.16 |
Adra1a |
adrenergic receptor, alpha 1a |
25111 |
0.21 |
| chr6_142451263_142451444 | 0.16 |
Gys2 |
glycogen synthase 2 |
21756 |
0.15 |
| chr1_140183261_140183634 | 0.16 |
Cfh |
complement component factor h |
36 |
0.98 |
| chr12_111435070_111435221 | 0.16 |
Gm10425 |
predicted gene 10425 |
4199 |
0.14 |
| chr7_28049499_28049670 | 0.16 |
Psmc4 |
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
464 |
0.71 |
| chr8_116872866_116873031 | 0.16 |
Cmc2 |
COX assembly mitochondrial protein 2 |
21576 |
0.18 |
| chr18_61374503_61374706 | 0.16 |
Gm25301 |
predicted gene, 25301 |
23209 |
0.12 |
| chr4_89162623_89162774 | 0.16 |
Gm12607 |
predicted gene 12607 |
507 |
0.72 |
| chr19_31896057_31896235 | 0.16 |
Gm19241 |
predicted gene, 19241 |
17498 |
0.2 |
| chr12_84034323_84034494 | 0.16 |
Acot4 |
acyl-CoA thioesterase 4 |
3971 |
0.1 |
| chr15_66942451_66942629 | 0.15 |
Ndrg1 |
N-myc downstream regulated gene 1 |
5939 |
0.18 |
| chr8_115980743_115980903 | 0.15 |
Gm45733 |
predicted gene 45733 |
8752 |
0.32 |
| chr8_115719399_115719579 | 0.15 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
11695 |
0.23 |
| chr1_184605964_184606231 | 0.15 |
Rpl21-ps1 |
ribosomal protein 21, pseudogene 1 |
11151 |
0.15 |
| chr17_88625924_88626075 | 0.15 |
Ston1 |
stonin 1 |
556 |
0.73 |
| chr18_75377563_75377744 | 0.15 |
Smad7 |
SMAD family member 7 |
2739 |
0.29 |
| chr18_20552765_20552968 | 0.15 |
Tpi-rs10 |
triosephosphate isomerase related sequence 10 |
3502 |
0.21 |
| chr7_100937600_100937760 | 0.15 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
5573 |
0.15 |
| chr1_31019080_31019348 | 0.15 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
19310 |
0.13 |
| chr8_122012297_122012477 | 0.15 |
Banp |
BTG3 associated nuclear protein |
11540 |
0.14 |
| chr14_63223276_63223490 | 0.15 |
Gata4 |
GATA binding protein 4 |
9776 |
0.16 |
| chr9_74790782_74790933 | 0.15 |
Gm22315 |
predicted gene, 22315 |
8787 |
0.19 |
| chr16_34083289_34083602 | 0.15 |
Kalrn |
kalirin, RhoGEF kinase |
12542 |
0.25 |
| chr18_16750214_16750554 | 0.15 |
Gm15485 |
predicted gene 15485 |
21651 |
0.21 |
| chr14_62329558_62329856 | 0.15 |
Rnaseh2b |
ribonuclease H2, subunit B |
2361 |
0.32 |
| chr2_122256732_122256883 | 0.15 |
Sord |
sorbitol dehydrogenase |
22058 |
0.09 |
| chr11_97920651_97920808 | 0.15 |
Gm22461 |
predicted gene, 22461 |
21011 |
0.1 |
| chr14_73116983_73117184 | 0.15 |
Rcbtb2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
5954 |
0.23 |
| chr11_80453320_80453471 | 0.15 |
Psmd11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
2973 |
0.25 |
| chr6_139839930_139840114 | 0.15 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
1423 |
0.45 |
| chr18_80275077_80275246 | 0.15 |
Slc66a2 |
solute carrier family 66 member 2 |
12321 |
0.11 |
| chr3_36854068_36854318 | 0.15 |
4932438A13Rik |
RIKEN cDNA 4932438A13 gene |
8911 |
0.21 |
| chr13_51149822_51150173 | 0.15 |
Spin1 |
spindlin 1 |
15678 |
0.17 |
| chr12_105686564_105686722 | 0.15 |
Gskip |
GSK3B interacting protein |
1242 |
0.32 |
| chr1_118458813_118459291 | 0.15 |
Gm22710 |
predicted gene, 22710 |
213 |
0.44 |
| chr9_114112478_114112646 | 0.15 |
AC157997.1 |
cartilage associated protein (Crtap) pseudogene |
4115 |
0.21 |
| chr18_35740040_35740191 | 0.15 |
Sting1 |
stimulator of interferon response cGAMP interactor 1 |
90 |
0.94 |
| chr5_91274508_91274681 | 0.14 |
Gm19619 |
predicted gene, 19619 |
8827 |
0.24 |
| chr17_32347830_32347981 | 0.14 |
Akap8l |
A kinase (PRKA) anchor protein 8-like |
2601 |
0.17 |
| chr2_34544709_34544892 | 0.14 |
Gm13408 |
predicted gene 13408 |
20199 |
0.2 |
| chr5_103848915_103849118 | 0.14 |
Klhl8 |
kelch-like 8 |
25353 |
0.14 |
| chr7_73426374_73426525 | 0.14 |
Gm44645 |
predicted gene 44645 |
4199 |
0.14 |
| chr3_96528686_96528855 | 0.14 |
Hjv |
hemojuvelin BMP co-receptor |
3598 |
0.09 |
| chr17_70847152_70847479 | 0.14 |
Tgif1 |
TGFB-induced factor homeobox 1 |
145 |
0.92 |
| chr9_74793041_74793363 | 0.14 |
Gm22315 |
predicted gene, 22315 |
11132 |
0.18 |
| chr8_3247329_3247509 | 0.14 |
Gm16180 |
predicted gene 16180 |
12285 |
0.19 |
| chr6_15728953_15729104 | 0.14 |
Mdfic |
MyoD family inhibitor domain containing |
1229 |
0.59 |
| chr1_194482743_194482912 | 0.14 |
Plxna2 |
plexin A2 |
135391 |
0.05 |
| chr1_91245290_91245453 | 0.14 |
Ube2f |
ubiquitin-conjugating enzyme E2F (putative) |
4933 |
0.17 |
| chr11_100935486_100935649 | 0.14 |
Stat3 |
signal transducer and activator of transcription 3 |
3813 |
0.17 |
| chr6_31198025_31198176 | 0.14 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
18266 |
0.14 |
| chr2_157819087_157819264 | 0.14 |
Ctnnbl1 |
catenin, beta like 1 |
29311 |
0.17 |
| chr11_106742585_106742746 | 0.14 |
Pecam1 |
platelet/endothelial cell adhesion molecule 1 |
7963 |
0.11 |
| chr16_43448600_43448866 | 0.14 |
Gm15713 |
predicted gene 15713 |
28537 |
0.18 |
| chr10_99314465_99314820 | 0.14 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
15476 |
0.14 |
| chr8_128481354_128481549 | 0.14 |
Nrp1 |
neuropilin 1 |
122054 |
0.05 |
| chr19_31896808_31896966 | 0.14 |
Gm19241 |
predicted gene, 19241 |
16757 |
0.2 |
| chr7_7281443_7281594 | 0.14 |
Gm45844 |
predicted gene 45844 |
3229 |
0.12 |
| chr2_65547147_65547392 | 0.14 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
17865 |
0.21 |
| chr3_95523824_95523997 | 0.14 |
Ctss |
cathepsin S |
2876 |
0.14 |
| chr16_31048299_31048452 | 0.14 |
Xxylt1 |
xyloside xylosyltransferase 1 |
13052 |
0.19 |
| chr19_55195901_55196052 | 0.14 |
Mir6715 |
microRNA 6715 |
3298 |
0.21 |
| chr5_75320369_75320658 | 0.14 |
Gm18345 |
predicted gene, 18345 |
21930 |
0.17 |
| chr9_114924578_114924744 | 0.14 |
Gpd1l |
glycerol-3-phosphate dehydrogenase 1-like |
5307 |
0.22 |
| chr10_59901049_59901200 | 0.14 |
Dnajb12 |
DnaJ heat shock protein family (Hsp40) member B12 |
21498 |
0.14 |
| chr2_60181625_60181942 | 0.14 |
Baz2b |
bromodomain adjacent to zinc finger domain, 2B |
27376 |
0.13 |
| chr6_8416975_8417147 | 0.14 |
Umad1 |
UMAP1-MVP12 associated (UMA) domain containing 1 |
2617 |
0.24 |
| chr5_34574967_34575118 | 0.14 |
Gm42867 |
predicted gene 42867 |
46 |
0.91 |
| chr10_86990632_86990973 | 0.14 |
Gm16269 |
predicted gene 16269 |
9495 |
0.15 |
| chr8_68020941_68021106 | 0.13 |
Gm22018 |
predicted gene, 22018 |
10683 |
0.24 |
| chr19_23024753_23025259 | 0.13 |
Gm50136 |
predicted gene, 50136 |
36448 |
0.17 |
| chr5_149160390_149160713 | 0.13 |
Gm8615 |
predicted pseudogene 8615 |
4342 |
0.1 |
| chr9_40330530_40330684 | 0.13 |
1700110K17Rik |
RIKEN cDNA 1700110K17 gene |
2867 |
0.15 |
| chr8_95800469_95800620 | 0.13 |
4930513N10Rik |
RIKEN cDNA 4930513N10 gene |
6200 |
0.09 |
| chr1_132384418_132384569 | 0.13 |
Gm15849 |
predicted gene 15849 |
3364 |
0.17 |
| chr8_11323805_11323964 | 0.13 |
Col4a1 |
collagen, type IV, alpha 1 |
11058 |
0.16 |
| chr8_109579164_109579323 | 0.13 |
Hp |
haptoglobin |
71 |
0.96 |
| chr3_97076477_97076637 | 0.13 |
4930573H18Rik |
RIKEN cDNA 4930573H18 gene |
16228 |
0.15 |
| chr9_113118608_113118982 | 0.13 |
Gm36251 |
predicted gene, 36251 |
4234 |
0.32 |
| chr8_26697737_26697895 | 0.13 |
Gm32098 |
predicted gene, 32098 |
29510 |
0.14 |
| chrX_169269868_169270019 | 0.13 |
Hccs |
holocytochrome c synthetase |
50337 |
0.13 |
| chr9_41938128_41938279 | 0.13 |
Sorl1 |
sortilin-related receptor, LDLR class A repeats-containing |
36384 |
0.13 |
| chr2_84996477_84996638 | 0.13 |
Prg3 |
proteoglycan 3 |
8342 |
0.12 |
| chrX_52140432_52140583 | 0.13 |
Gpc4 |
glypican 4 |
24745 |
0.24 |
| chr4_130774522_130774687 | 0.13 |
Gm23711 |
predicted gene, 23711 |
1137 |
0.34 |
| chr10_58370276_58370427 | 0.13 |
Lims1 |
LIM and senescent cell antigen-like domains 1 |
1103 |
0.51 |
| chr1_83900189_83900548 | 0.13 |
4933436I20Rik |
RIKEN cDNA 4933436I20 gene |
11203 |
0.23 |
| chr16_95383459_95383635 | 0.13 |
Gm31641 |
predicted gene, 31641 |
27871 |
0.2 |
| chr6_128503302_128503638 | 0.12 |
Pzp |
PZP, alpha-2-macroglobulin like |
8247 |
0.09 |
| chr4_150434207_150434380 | 0.12 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
27899 |
0.2 |
| chr19_32627970_32628365 | 0.12 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
8162 |
0.23 |
| chr10_117677088_117677246 | 0.12 |
Cpm |
carboxypeptidase M |
1274 |
0.36 |
| chr2_128266444_128266595 | 0.12 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
25834 |
0.2 |
| chr1_152573046_152573282 | 0.12 |
Rgl1 |
ral guanine nucleotide dissociation stimulator,-like 1 |
19595 |
0.23 |
| chr9_119983098_119983399 | 0.12 |
1700019L13Rik |
RIKEN cDNA 1700019L13 gene |
31 |
0.73 |
| chr2_131330687_131330851 | 0.12 |
Rnf24 |
ring finger protein 24 |
1781 |
0.25 |
| chr5_143492763_143492920 | 0.12 |
Daglb |
diacylglycerol lipase, beta |
6163 |
0.13 |
| chr11_61273784_61273967 | 0.12 |
Aldh3a2 |
aldehyde dehydrogenase family 3, subfamily A2 |
6411 |
0.17 |
| chr8_8959238_8959490 | 0.12 |
Gm44515 |
predicted gene 44515 |
26665 |
0.21 |
| chr16_23144476_23144635 | 0.12 |
Adipoq |
adiponectin, C1Q and collagen domain containing |
1981 |
0.13 |
| chr9_116847373_116847540 | 0.12 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
24637 |
0.27 |
| chr4_118033071_118033392 | 0.12 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
1349 |
0.44 |
| chr4_35101216_35101367 | 0.12 |
Ifnk |
interferon kappa |
50765 |
0.12 |
| chr19_55808091_55808481 | 0.12 |
Ppnr |
per-pentamer repeat gene |
33386 |
0.21 |
| chr16_97281118_97281506 | 0.12 |
Bace2 |
beta-site APP-cleaving enzyme 2 |
75430 |
0.1 |
| chr7_65802914_65803076 | 0.12 |
1810008I18Rik |
RIKEN cDNA 1810008I18 gene |
116 |
0.96 |
| chr3_10007581_10007909 | 0.12 |
Fabp5 |
fatty acid binding protein 5, epidermal |
4803 |
0.19 |
| chr6_70814231_70814401 | 0.12 |
Rpia |
ribose 5-phosphate isomerase A |
22084 |
0.13 |
| chr16_34826235_34826441 | 0.12 |
Mylk |
myosin, light polypeptide kinase |
41417 |
0.17 |
| chr17_34896533_34896711 | 0.12 |
C2 |
complement component 2 (within H-2S) |
1065 |
0.19 |
| chr13_98917969_98918176 | 0.12 |
Tnpo1 |
transportin 1 |
8282 |
0.14 |
| chr8_110822079_110822262 | 0.12 |
Il34 |
interleukin 34 |
16246 |
0.1 |
| chr7_121836969_121837120 | 0.12 |
Gm44740 |
predicted gene 44740 |
2021 |
0.31 |
| chr15_27782797_27782987 | 0.12 |
Trio |
triple functional domain (PTPRF interacting) |
5746 |
0.25 |
| chr1_160980118_160980282 | 0.12 |
Serpinc1 |
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
1512 |
0.18 |
| chr4_55354398_55354553 | 0.12 |
Rad23b |
RAD23 homolog B, nucleotide excision repair protein |
4385 |
0.18 |
| chr16_6039608_6039759 | 0.11 |
1700123O21Rik |
RIKEN cDNA 1700123O21 gene |
64098 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0003284 | septum primum development(GO:0003284) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0052622 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |