Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
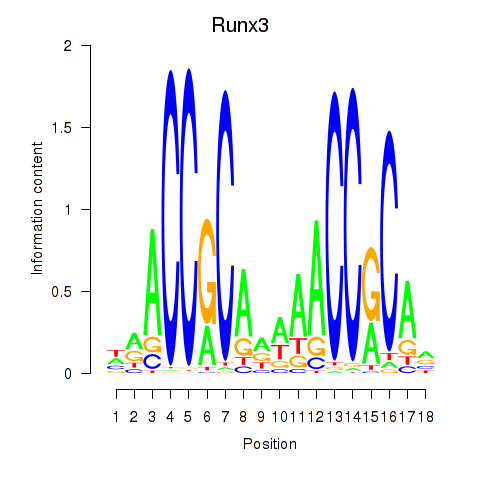
Results for Runx3
Z-value: 2.00
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSMUSG00000070691.4 | runt related transcription factor 3 |
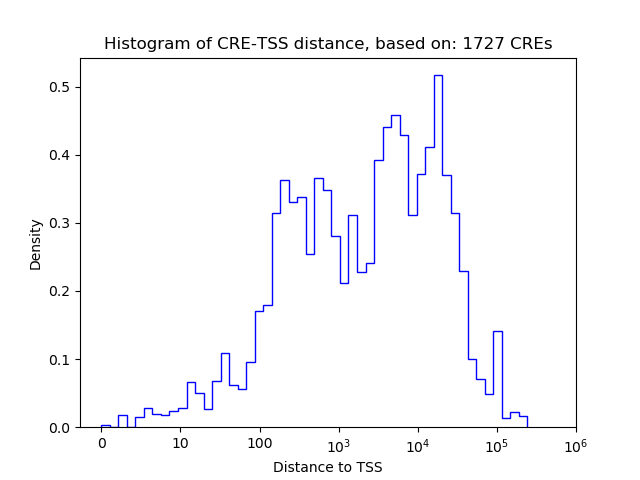
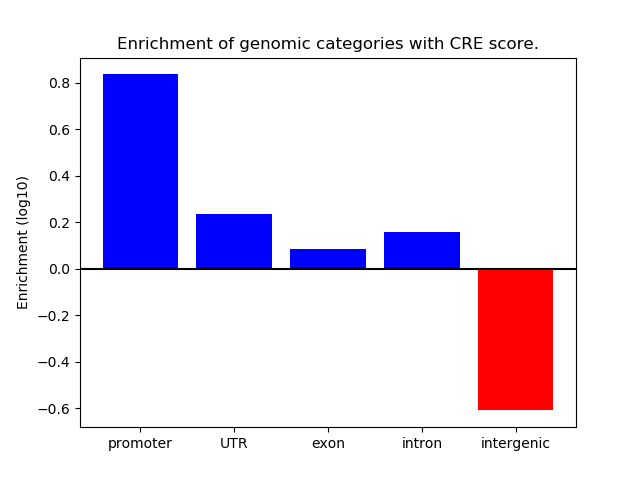
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_135204278_135204439 | Runx3 | 49922 | 0.096326 | 0.75 | 8.8e-02 | Click! |
| chr4_135203380_135204093 | Runx3 | 49300 | 0.097535 | 0.58 | 2.3e-01 | Click! |
| chr4_135206185_135206602 | Runx3 | 51957 | 0.092384 | 0.56 | 2.5e-01 | Click! |
Activity of the Runx3 motif across conditions
Conditions sorted by the z-value of the Runx3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
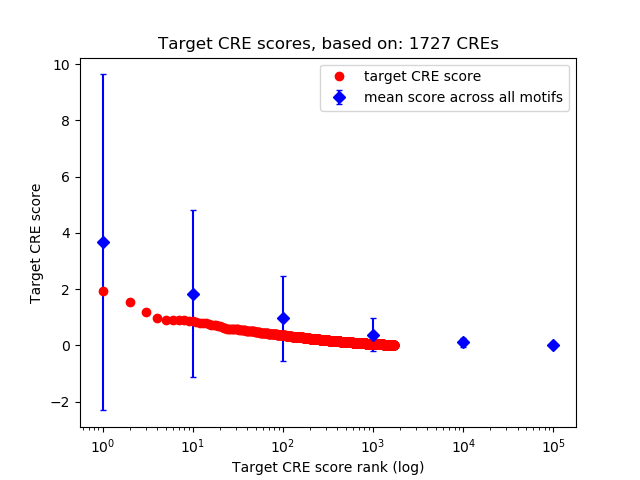
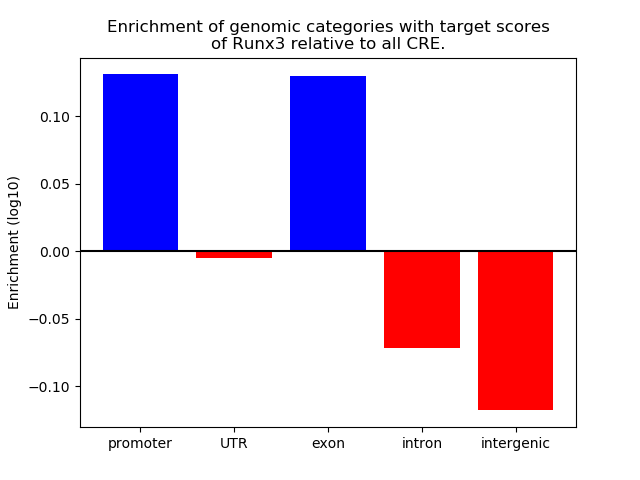
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_109993740_109993939 | 1.94 |
Gm34667 |
predicted gene, 34667 |
30034 |
0.1 |
| chr16_21613201_21613352 | 1.53 |
Vps8 |
VPS8 CORVET complex subunit |
5430 |
0.27 |
| chr12_41247016_41247167 | 1.17 |
Gm47376 |
predicted gene, 47376 |
100999 |
0.07 |
| chr15_102192910_102193280 | 0.98 |
Csad |
cysteine sulfinic acid decarboxylase |
1036 |
0.35 |
| chr9_103066985_103067146 | 0.92 |
Gm47468 |
predicted gene, 47468 |
18461 |
0.14 |
| chr10_7976561_7976927 | 0.91 |
Tab2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
20514 |
0.19 |
| chr13_93621073_93621244 | 0.90 |
Gm15622 |
predicted gene 15622 |
4224 |
0.19 |
| chr3_123104065_123104239 | 0.90 |
Synpo2 |
synaptopodin 2 |
8658 |
0.16 |
| chr10_95544966_95545117 | 0.88 |
Ube2n |
ubiquitin-conjugating enzyme E2N |
3528 |
0.16 |
| chr2_77155937_77156088 | 0.88 |
Ccdc141 |
coiled-coil domain containing 141 |
14564 |
0.21 |
| chr15_99093772_99093973 | 0.81 |
Dnajc22 |
DnaJ heat shock protein family (Hsp40) member C22 |
702 |
0.47 |
| chr6_88717841_88718269 | 0.80 |
Mgll |
monoglyceride lipase |
6357 |
0.14 |
| chr2_103600397_103600730 | 0.80 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
34253 |
0.15 |
| chr7_81542702_81542853 | 0.80 |
Gm37829 |
predicted gene, 37829 |
10256 |
0.1 |
| chr4_86664611_86664778 | 0.74 |
Plin2 |
perilipin 2 |
1031 |
0.52 |
| chr6_85803570_85803735 | 0.74 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
6211 |
0.09 |
| chr5_9103732_9103883 | 0.72 |
Tmem243 |
transmembrane protein 243, mitochondrial |
3060 |
0.23 |
| chr7_80666924_80667352 | 0.72 |
Crtc3 |
CREB regulated transcription coactivator 3 |
21739 |
0.15 |
| chr6_85702238_85702435 | 0.70 |
Nat8f7 |
N-acetyltransferase 8 (GCN5-related) family member 7 |
5522 |
0.12 |
| chr4_46657221_46657388 | 0.67 |
Tbc1d2 |
TBC1 domain family, member 2 |
7095 |
0.21 |
| chr16_50764320_50764471 | 0.65 |
1700116B05Rik |
RIKEN cDNA 1700116B05 gene |
19014 |
0.18 |
| chr15_82455207_82455652 | 0.60 |
Cyp2d9 |
cytochrome P450, family 2, subfamily d, polypeptide 9 |
141 |
0.87 |
| chr5_120566729_120566934 | 0.60 |
2510016D11Rik |
RIKEN cDNA 2510016D11 gene |
13792 |
0.09 |
| chr2_161090855_161091012 | 0.59 |
Chd6 |
chromodomain helicase DNA binding protein 6 |
18071 |
0.18 |
| chr7_35802685_35802845 | 0.59 |
E130304I02Rik |
RIKEN cDNA E130304I02 gene |
172 |
0.52 |
| chr4_100401449_100401626 | 0.58 |
Gm12706 |
predicted gene 12706 |
39478 |
0.18 |
| chr15_82397705_82398277 | 0.58 |
Cyp2d11 |
cytochrome P450, family 2, subfamily d, polypeptide 11 |
3969 |
0.07 |
| chr1_39545677_39545839 | 0.57 |
Snord89 |
small nucleolar RNA, C/D box 89 |
3096 |
0.14 |
| chr18_20980846_20980997 | 0.57 |
Rnf125 |
ring finger protein 125 |
19449 |
0.18 |
| chr11_67526441_67526592 | 0.57 |
Gas7 |
growth arrest specific 7 |
20638 |
0.21 |
| chr8_71535881_71536032 | 0.57 |
Bst2 |
bone marrow stromal cell antigen 2 |
1375 |
0.2 |
| chr2_172477517_172477864 | 0.57 |
Fam209 |
family with sequence similarity 209 |
5170 |
0.15 |
| chr16_17885845_17885996 | 0.56 |
Dgcr2 |
DiGeorge syndrome critical region gene 2 |
5673 |
0.09 |
| chr18_63597933_63598104 | 0.56 |
Gm24848 |
predicted gene, 24848 |
13282 |
0.18 |
| chr5_125137828_125138031 | 0.55 |
Ncor2 |
nuclear receptor co-repressor 2 |
13029 |
0.2 |
| chr11_70409620_70409854 | 0.53 |
Pelp1 |
proline, glutamic acid and leucine rich protein 1 |
291 |
0.78 |
| chr4_101416398_101416601 | 0.53 |
Ak4 |
adenylate kinase 4 |
2778 |
0.2 |
| chr7_49633783_49633973 | 0.53 |
Dbx1 |
developing brain homeobox 1 |
2971 |
0.33 |
| chr1_182602082_182602275 | 0.53 |
Capn8 |
calpain 8 |
37140 |
0.13 |
| chr15_80711397_80711548 | 0.52 |
Tnrc6b |
trinucleotide repeat containing 6b |
159 |
0.88 |
| chr4_155320479_155320646 | 0.51 |
Prkcz |
protein kinase C, zeta |
19391 |
0.14 |
| chr15_10953321_10953904 | 0.51 |
C1qtnf3 |
C1q and tumor necrosis factor related protein 3 |
1255 |
0.4 |
| chr1_193617502_193617686 | 0.51 |
Mir205hg |
Mir205 host gene |
107420 |
0.06 |
| chr2_126574664_126574815 | 0.50 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
9979 |
0.18 |
| chr17_24470568_24470947 | 0.50 |
Pgp |
phosphoglycolate phosphatase |
322 |
0.51 |
| chr6_5190649_5191077 | 0.50 |
Pon1 |
paraoxonase 1 |
2900 |
0.26 |
| chr15_36471981_36472313 | 0.50 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr8_109705135_109705296 | 0.49 |
Zfp821 |
zinc finger protein 821 |
331 |
0.85 |
| chr5_135090387_135090722 | 0.49 |
Mlxipl |
MLX interacting protein-like |
664 |
0.5 |
| chr12_26398692_26398891 | 0.48 |
Rnf144a |
ring finger protein 144A |
7656 |
0.16 |
| chr13_98946494_98946645 | 0.48 |
Gm35215 |
predicted gene, 35215 |
653 |
0.6 |
| chr14_73630476_73630939 | 0.47 |
Gm21750 |
predicted gene, 21750 |
30518 |
0.15 |
| chr13_119966643_119967735 | 0.47 |
Gm20784 |
predicted gene, 20784 |
4945 |
0.11 |
| chr9_48826985_48827136 | 0.47 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
8885 |
0.24 |
| chr11_60728592_60728763 | 0.46 |
Mir5100 |
microRNA 5100 |
14 |
0.73 |
| chr11_69902083_69902235 | 0.45 |
Neurl4 |
neuralized E3 ubiquitin protein ligase 4 |
3 |
0.9 |
| chr12_98628262_98628451 | 0.45 |
Spata7 |
spermatogenesis associated 7 |
159 |
0.93 |
| chr7_130710473_130710636 | 0.45 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
3123 |
0.24 |
| chr2_69226745_69226961 | 0.45 |
G6pc2 |
glucose-6-phosphatase, catalytic, 2 |
6825 |
0.15 |
| chr3_22168181_22168357 | 0.44 |
Tbl1xr1 |
transducin (beta)-like 1X-linked receptor 1 |
19327 |
0.21 |
| chr11_75678792_75678954 | 0.44 |
Crk |
v-crk avian sarcoma virus CT10 oncogene homolog |
386 |
0.8 |
| chr17_27556354_27556505 | 0.44 |
Hmga1 |
high mobility group AT-hook 1 |
68 |
0.51 |
| chr14_69171500_69171815 | 0.43 |
Nkx2-6 |
NK2 homeobox 6 |
145 |
0.57 |
| chr2_72475881_72476264 | 0.43 |
Cdca7 |
cell division cycle associated 7 |
87 |
0.98 |
| chr12_55487707_55488022 | 0.43 |
Nfkbia |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
3946 |
0.21 |
| chr12_87300349_87300500 | 0.42 |
Ism2 |
isthmin 2 |
719 |
0.52 |
| chr11_120555335_120555502 | 0.42 |
Ppp1r27 |
protein phosphatase 1, regulatory subunit 27 |
4286 |
0.07 |
| chr5_132467140_132467305 | 0.42 |
Auts2 |
autism susceptibility candidate 2 |
17862 |
0.12 |
| chr3_149546578_149546753 | 0.42 |
Gm30382 |
predicted gene, 30382 |
101364 |
0.08 |
| chr5_145852474_145852695 | 0.42 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
24107 |
0.13 |
| chr9_124281840_124282030 | 0.41 |
Gm39469 |
predicted gene, 39469 |
204 |
0.93 |
| chr8_105347302_105347515 | 0.41 |
Fhod1 |
formin homology 2 domain containing 1 |
501 |
0.52 |
| chr2_132263417_132263641 | 0.41 |
Cds2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
288 |
0.88 |
| chrX_74305335_74305495 | 0.40 |
Gdi1 |
guanosine diphosphate (GDP) dissociation inhibitor 1 |
102 |
0.89 |
| chr12_26469202_26469522 | 0.40 |
Cmpk2 |
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
158 |
0.94 |
| chr15_75974567_75974718 | 0.40 |
Zfp707 |
zinc finger protein 707 |
1859 |
0.15 |
| chr8_61574352_61574983 | 0.40 |
Palld |
palladin, cytoskeletal associated protein |
16472 |
0.25 |
| chr2_118490490_118490877 | 0.40 |
Gm13983 |
predicted gene 13983 |
6506 |
0.16 |
| chr4_41713984_41714147 | 0.40 |
Rpp25l |
ribonuclease P/MRP 25 subunit-like |
531 |
0.59 |
| chr2_4651833_4652008 | 0.40 |
Prpf18 |
pre-mRNA processing factor 18 |
161 |
0.96 |
| chr5_76184387_76184581 | 0.40 |
Tmem165 |
transmembrane protein 165 |
534 |
0.73 |
| chr13_33003844_33004060 | 0.40 |
Serpinb9 |
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
589 |
0.64 |
| chr2_154855271_154855679 | 0.39 |
Eif2s2 |
eukaryotic translation initiation factor 2, subunit 2 (beta) |
37307 |
0.13 |
| chr3_58593720_58593871 | 0.39 |
Selenot |
selenoprotein T |
17097 |
0.14 |
| chr8_48492721_48492872 | 0.39 |
Tenm3 |
teneurin transmembrane protein 3 |
62517 |
0.14 |
| chr5_112282209_112282362 | 0.38 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
5578 |
0.13 |
| chr16_91238840_91239051 | 0.38 |
Olig2 |
oligodendrocyte transcription factor 2 |
13488 |
0.11 |
| chr9_108792795_108793094 | 0.38 |
Gm23034 |
predicted gene, 23034 |
2444 |
0.14 |
| chr19_41846454_41846665 | 0.37 |
Frat2 |
frequently rearranged in advanced T cell lymphomas 2 |
1573 |
0.3 |
| chrX_140568088_140568285 | 0.37 |
AL683809.1 |
TSC22 domain family, member 3 (Tsc22d3), pseuodgene |
15413 |
0.17 |
| chr4_12140361_12140643 | 0.37 |
Rbm12b1 |
RNA binding motif protein 12 B1 |
185 |
0.91 |
| chr6_86820856_86821008 | 0.37 |
Anxa4 |
annexin A4 |
27348 |
0.1 |
| chr11_83853555_83853722 | 0.37 |
Hnf1b |
HNF1 homeobox B |
678 |
0.62 |
| chr7_43444450_43444615 | 0.36 |
Etfb |
electron transferring flavoprotein, beta polypeptide |
382 |
0.4 |
| chr8_70131574_70131799 | 0.36 |
Nr2c2ap |
nuclear receptor 2C2-associated protein |
339 |
0.73 |
| chr8_109869084_109869235 | 0.36 |
Phlpp2 |
PH domain and leucine rich repeat protein phosphatase 2 |
548 |
0.48 |
| chr5_125323526_125323720 | 0.36 |
Gm42633 |
predicted gene 42633 |
2179 |
0.22 |
| chr8_121307859_121308016 | 0.36 |
Gm26815 |
predicted gene, 26815 |
48899 |
0.13 |
| chr15_99034485_99034683 | 0.36 |
Tuba1c |
tubulin, alpha 1C |
4263 |
0.11 |
| chr10_44527174_44527473 | 0.36 |
Prdm1 |
PR domain containing 1, with ZNF domain |
1178 |
0.46 |
| chr10_53928699_53928881 | 0.36 |
Man1a |
mannosidase 1, alpha |
48325 |
0.16 |
| chr3_94399233_94399394 | 0.35 |
Lingo4 |
leucine rich repeat and Ig domain containing 4 |
796 |
0.33 |
| chr18_50137858_50138009 | 0.35 |
Cd63-ps |
CD63 antigen, pseudogene |
3138 |
0.23 |
| chr14_34618565_34618885 | 0.35 |
Opn4 |
opsin 4 (melanopsin) |
18583 |
0.11 |
| chr14_61354089_61354266 | 0.35 |
Ebpl |
emopamil binding protein-like |
6143 |
0.13 |
| chr11_20733626_20734042 | 0.35 |
Gm12036 |
predicted gene 12036 |
4414 |
0.21 |
| chr12_30200282_30200708 | 0.34 |
Sntg2 |
syntrophin, gamma 2 |
840 |
0.72 |
| chr5_35030633_35030784 | 0.34 |
Gm43791 |
predicted gene 43791 |
2737 |
0.18 |
| chr10_7211859_7212010 | 0.34 |
Cnksr3 |
Cnksr family member 3 |
303 |
0.94 |
| chr4_48046634_48046887 | 0.33 |
Nr4a3 |
nuclear receptor subfamily 4, group A, member 3 |
1529 |
0.46 |
| chr4_134879692_134879843 | 0.33 |
Rhd |
Rh blood group, D antigen |
43 |
0.97 |
| chr12_113221349_113221516 | 0.33 |
Gm25622 |
predicted gene, 25622 |
7283 |
0.14 |
| chr4_100129098_100129249 | 0.33 |
Gm12701 |
predicted gene 12701 |
9552 |
0.23 |
| chr10_122986042_122986255 | 0.32 |
D630033A02Rik |
RIKEN cDNA D630033A02 gene |
317 |
0.58 |
| chr4_12089474_12090115 | 0.32 |
Tmem67 |
transmembrane protein 67 |
226 |
0.62 |
| chr4_135270319_135270473 | 0.32 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
2418 |
0.22 |
| chr10_127513114_127513548 | 0.32 |
Ndufa4l2 |
Ndufa4, mitochondrial complex associated like 2 |
1636 |
0.21 |
| chr17_30199906_30200207 | 0.32 |
Zfand3 |
zinc finger, AN1-type domain 3 |
2480 |
0.27 |
| chr8_33930013_33930185 | 0.32 |
Rbpms |
RNA binding protein gene with multiple splicing |
236 |
0.92 |
| chr10_25621751_25621928 | 0.32 |
Gm36908 |
predicted gene, 36908 |
276 |
0.91 |
| chr19_41847242_41847459 | 0.32 |
Frat2 |
frequently rearranged in advanced T cell lymphomas 2 |
782 |
0.55 |
| chr11_120086018_120086485 | 0.32 |
Cep131 |
centrosomal protein 131 |
576 |
0.57 |
| chr11_109718016_109718204 | 0.32 |
Fam20a |
family with sequence similarity 20, member A |
4146 |
0.22 |
| chr4_32240053_32240204 | 0.32 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
1051 |
0.54 |
| chr12_84939224_84939386 | 0.32 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
4073 |
0.15 |
| chr3_96457950_96458248 | 0.31 |
Gm10685 |
predicted gene 10685 |
110 |
0.87 |
| chr3_138422238_138422402 | 0.31 |
Adh4 |
alcohol dehydrogenase 4 (class II), pi polypeptide |
6825 |
0.14 |
| chr1_184785241_184785692 | 0.31 |
Mtarc1 |
mitochondrial amidoxime reducing component 1 |
23387 |
0.12 |
| chr15_46598446_46598597 | 0.31 |
4930548G14Rik |
RIKEN cDNA 4930548G14 gene |
24784 |
0.25 |
| chr13_59004016_59004334 | 0.31 |
Gm34245 |
predicted gene, 34245 |
74121 |
0.08 |
| chr4_59311382_59311545 | 0.31 |
Susd1 |
sushi domain containing 1 |
4503 |
0.24 |
| chr16_91362482_91362649 | 0.31 |
Ifnar2 |
interferon (alpha and beta) receptor 2 |
10218 |
0.11 |
| chr4_132535706_132535867 | 0.31 |
Dnajc8 |
DnaJ heat shock protein family (Hsp40) member C8 |
127 |
0.91 |
| chr3_96452393_96452678 | 0.31 |
BC107364 |
cDNA sequence BC107364 |
229 |
0.76 |
| chr11_120466994_120467145 | 0.31 |
Arl16 |
ADP-ribosylation factor-like 16 |
205 |
0.71 |
| chr10_81135573_81135724 | 0.31 |
Zbtb7a |
zinc finger and BTB domain containing 7a |
428 |
0.63 |
| chr2_60383071_60383222 | 0.31 |
Ly75 |
lymphocyte antigen 75 |
85 |
0.97 |
| chr13_60665524_60665709 | 0.30 |
Gm48583 |
predicted gene, 48583 |
23826 |
0.15 |
| chr5_142782966_142783117 | 0.30 |
Tnrc18 |
trinucleotide repeat containing 18 |
5482 |
0.21 |
| chr4_128981174_128981543 | 0.30 |
Ak2 |
adenylate kinase 2 |
10600 |
0.15 |
| chr17_25507894_25508055 | 0.30 |
Gm33727 |
predicted gene, 33727 |
10611 |
0.09 |
| chr8_93227828_93228167 | 0.30 |
Ces1e |
carboxylesterase 1E |
1621 |
0.29 |
| chr8_85007833_85007986 | 0.30 |
Best2 |
bestrophin 2 |
5682 |
0.06 |
| chr4_76343359_76343684 | 0.30 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
722 |
0.77 |
| chr5_43781403_43781714 | 0.30 |
Fbxl5 |
F-box and leucine-rich repeat protein 5 |
542 |
0.46 |
| chr14_63248471_63248829 | 0.30 |
Gata4 |
GATA binding protein 4 |
3379 |
0.22 |
| chrX_13845208_13845423 | 0.30 |
Cask |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
773 |
0.75 |
| chr11_20561304_20561596 | 0.30 |
Gm12035 |
predicted gene 12035 |
13739 |
0.2 |
| chr10_30200517_30200678 | 0.30 |
Cenpw |
centromere protein W |
37 |
0.98 |
| chr19_10239849_10240587 | 0.29 |
Myrf |
myelin regulatory factor |
386 |
0.77 |
| chr6_17694870_17695095 | 0.29 |
St7 |
suppression of tumorigenicity 7 |
815 |
0.54 |
| chr7_31053117_31053317 | 0.29 |
Fxyd1 |
FXYD domain-containing ion transport regulator 1 |
95 |
0.91 |
| chr17_56104393_56104561 | 0.29 |
Hdgfl2 |
HDGF like 2 |
4915 |
0.09 |
| chr19_23105417_23105568 | 0.29 |
2410080I02Rik |
RIKEN cDNA 2410080I02 gene |
29408 |
0.13 |
| chr5_102068968_102069164 | 0.29 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
855 |
0.46 |
| chr17_83897414_83897589 | 0.29 |
1810073O08Rik |
RIKEN cDNA 1810073O08 gene |
20436 |
0.13 |
| chr5_100673907_100674085 | 0.28 |
Coq2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
160 |
0.94 |
| chr16_59594968_59595140 | 0.28 |
Crybg3 |
beta-gamma crystallin domain containing 3 |
5925 |
0.21 |
| chr12_59235308_59235463 | 0.28 |
Gm27284 |
predicted gene, 27284 |
1791 |
0.28 |
| chr3_107631354_107631565 | 0.28 |
Gm10961 |
predicted gene 10961 |
137 |
0.63 |
| chr2_103472788_103473367 | 0.28 |
Cat |
catalase |
12048 |
0.18 |
| chr2_26922374_26922531 | 0.28 |
Surf2 |
surfeit gene 2 |
5024 |
0.08 |
| chr15_76606976_76607461 | 0.28 |
Cpsf1 |
cleavage and polyadenylation specific factor 1 |
271 |
0.77 |
| chr1_131971555_131972013 | 0.28 |
Slc45a3 |
solute carrier family 45, member 3 |
1175 |
0.35 |
| chr6_113494556_113494760 | 0.28 |
Creld1 |
cysteine-rich with EGF-like domains 1 |
2639 |
0.11 |
| chr7_132852781_132852967 | 0.28 |
Eef1akmt2 |
EEF1A lysine methyltransferase 2 |
201 |
0.91 |
| chr10_89770056_89770207 | 0.28 |
Uhrf1bp1l |
UHRF1 (ICBP90) binding protein 1-like |
17069 |
0.18 |
| chr9_61914762_61915200 | 0.28 |
Rplp1 |
ribosomal protein, large, P1 |
439 |
0.83 |
| chr8_27054619_27054804 | 0.27 |
Plpbp |
pyridoxal phosphate binding protein |
3937 |
0.13 |
| chr1_139422425_139422762 | 0.27 |
Zbtb41 |
zinc finger and BTB domain containing 41 |
211 |
0.92 |
| chr7_110942462_110942613 | 0.27 |
Mrvi1 |
MRV integration site 1 |
3650 |
0.23 |
| chr14_20995157_20995320 | 0.27 |
Vcl |
vinculin |
26765 |
0.17 |
| chr10_80702834_80702996 | 0.27 |
Izumo4 |
IZUMO family member 4 |
144 |
0.87 |
| chr11_116187883_116188093 | 0.27 |
Acox1 |
acyl-Coenzyme A oxidase 1, palmitoyl |
1587 |
0.21 |
| chr4_145774583_145774829 | 0.27 |
Gm13243 |
predicted gene 13243 |
13 |
0.94 |
| chr1_74723320_74723471 | 0.27 |
Cyp27a1 |
cytochrome P450, family 27, subfamily a, polypeptide 1 |
9821 |
0.12 |
| chr9_63550824_63550989 | 0.27 |
Gm16759 |
predicted gene, 16759 |
19554 |
0.18 |
| chr7_29155799_29155976 | 0.27 |
Fam98c |
family with sequence similarity 98, member C |
168 |
0.52 |
| chr7_19095153_19095353 | 0.27 |
Six5 |
sine oculis-related homeobox 5 |
659 |
0.44 |
| chr1_74016328_74016481 | 0.27 |
Tns1 |
tensin 1 |
18 |
0.98 |
| chr2_103595080_103595270 | 0.27 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
28865 |
0.16 |
| chr17_44256731_44256898 | 0.27 |
Clic5 |
chloride intracellular channel 5 |
19708 |
0.26 |
| chr2_153632897_153633072 | 0.27 |
Commd7 |
COMM domain containing 7 |
203 |
0.93 |
| chr1_93128050_93128201 | 0.26 |
Gm28086 |
predicted gene 28086 |
4022 |
0.15 |
| chr9_96718101_96718252 | 0.26 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
1389 |
0.38 |
| chr2_92376664_92376833 | 0.26 |
Pex16 |
peroxisomal biogenesis factor 16 |
1189 |
0.28 |
| chr6_66896024_66896206 | 0.26 |
Gng12 |
guanine nucleotide binding protein (G protein), gamma 12 |
282 |
0.6 |
| chr6_141327961_141328118 | 0.26 |
Gm10400 |
predicted gene 10400 |
12514 |
0.24 |
| chr7_132811347_132811663 | 0.25 |
Fam53b |
family with sequence similarity 53, member B |
1590 |
0.34 |
| chr16_24629324_24629735 | 0.25 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
92326 |
0.08 |
| chr17_24349257_24349433 | 0.25 |
Abca17 |
ATP-binding cassette, sub-family A (ABC1), member 17 |
1684 |
0.2 |
| chr10_28387713_28387881 | 0.25 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
172354 |
0.03 |
| chr1_90238643_90238822 | 0.25 |
Gm38277 |
predicted gene, 38277 |
10206 |
0.16 |
| chr3_8961467_8961864 | 0.25 |
Tpd52 |
tumor protein D52 |
588 |
0.76 |
| chr16_15888920_15889238 | 0.25 |
Cebpd |
CCAAT/enhancer binding protein (C/EBP), delta |
663 |
0.71 |
| chr5_125389673_125389844 | 0.25 |
Ubc |
ubiquitin C |
256 |
0.64 |
| chr9_62355457_62355637 | 0.25 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
10748 |
0.2 |
| chr11_60727211_60727861 | 0.24 |
Flii |
flightless I actin binding protein |
273 |
0.67 |
| chr10_93432561_93432735 | 0.24 |
Mir7688 |
microRNA 7688 |
459 |
0.78 |
| chr5_38260278_38260440 | 0.24 |
Tmem128 |
transmembrane protein 128 |
12 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.4 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.3 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.3 | GO:0010255 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.1 | 0.4 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.2 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.3 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.2 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.2 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.2 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.0 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.8 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0061197 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.0 | GO:0072197 | ureter morphogenesis(GO:0072197) metanephric nephron tubule formation(GO:0072289) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.0 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0034944 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0051735 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |