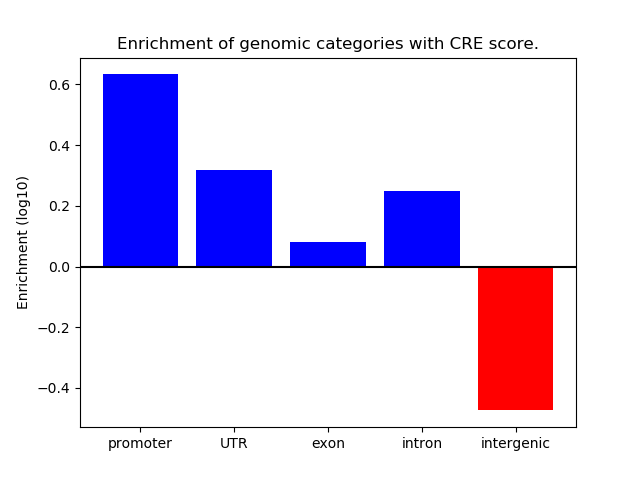
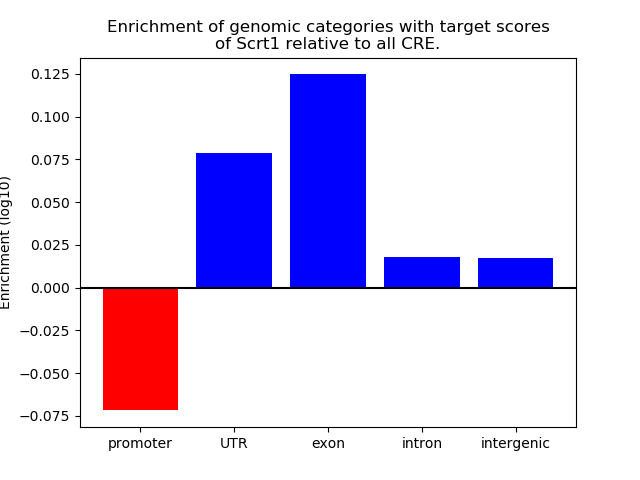
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
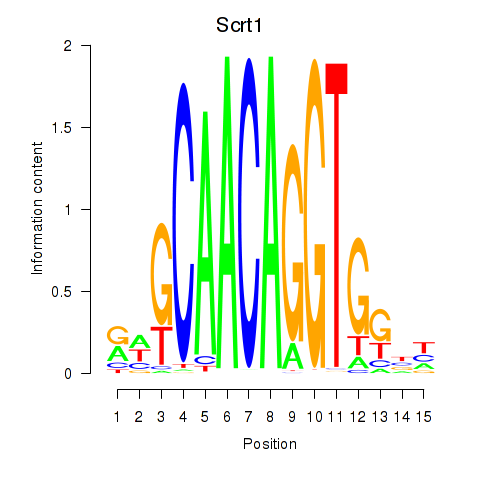
Results for Scrt1
Z-value: 0.89
Transcription factors associated with Scrt1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt1
|
ENSMUSG00000048385.8 | scratch family zinc finger 1 |
Activity of the Scrt1 motif across conditions
Conditions sorted by the z-value of the Scrt1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
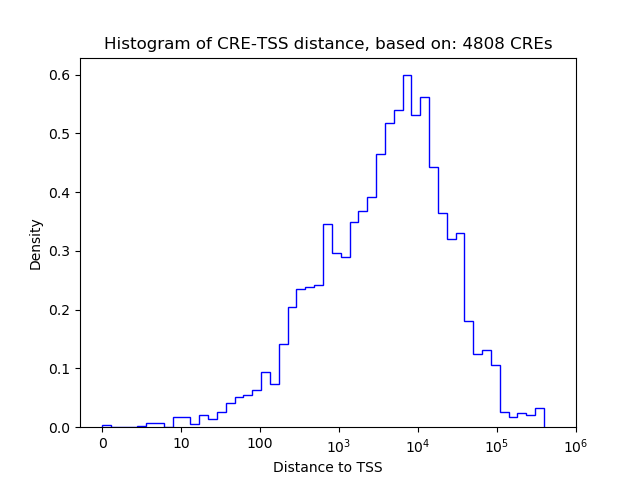
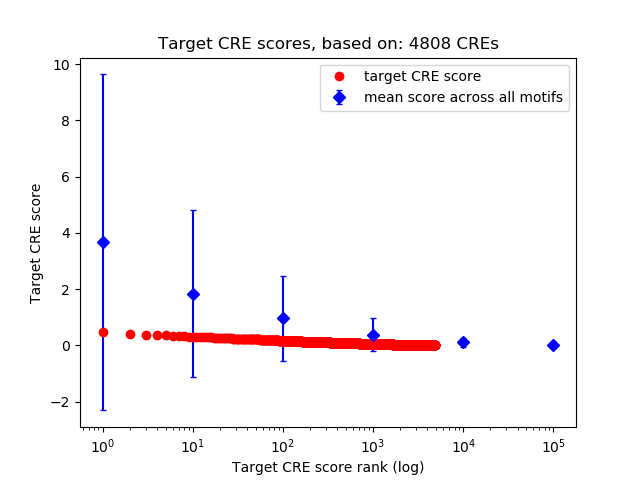
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_34073715_34073878 | 0.48 |
Tubb2a |
tubulin, beta 2A class IIA |
4211 |
0.12 |
| chr6_121890040_121890777 | 0.39 |
Mug1 |
murinoglobulin 1 |
4831 |
0.21 |
| chr2_71633995_71634167 | 0.38 |
Platr26 |
pluripotency associated transcript 26 |
85336 |
0.06 |
| chr3_83005590_83005767 | 0.36 |
Fgg |
fibrinogen gamma chain |
2046 |
0.24 |
| chr11_49088297_49088883 | 0.36 |
Gm12188 |
predicted gene 12188 |
47 |
0.79 |
| chr11_116108287_116108624 | 0.35 |
Trim47 |
tripartite motif-containing 47 |
336 |
0.77 |
| chr18_46718177_46718358 | 0.34 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
9762 |
0.13 |
| chr5_8985691_8985842 | 0.32 |
Crot |
carnitine O-octanoyltransferase |
3622 |
0.15 |
| chr1_183297724_183297875 | 0.31 |
Aida |
axin interactor, dorsalization associated |
229 |
0.74 |
| chr5_89033271_89033463 | 0.31 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
5275 |
0.32 |
| chr17_28272441_28272592 | 0.31 |
Ppard |
peroxisome proliferator activator receptor delta |
397 |
0.76 |
| chr3_135582591_135582742 | 0.30 |
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
11775 |
0.16 |
| chr11_102217522_102217681 | 0.30 |
Hdac5 |
histone deacetylase 5 |
1327 |
0.26 |
| chr12_109988471_109988667 | 0.29 |
Gm34667 |
predicted gene, 34667 |
35304 |
0.09 |
| chr13_55563371_55563565 | 0.29 |
Fam193b |
family with sequence similarity 193, member B |
4560 |
0.1 |
| chr11_82573919_82574081 | 0.28 |
Gm24612 |
predicted gene, 24612 |
22179 |
0.19 |
| chr6_57417438_57417602 | 0.27 |
Vmn1r20 |
vomeronasal 1 receptor 20 |
11410 |
0.12 |
| chr18_20936854_20937178 | 0.27 |
Rnf125 |
ring finger protein 125 |
7609 |
0.22 |
| chr12_21141829_21142079 | 0.27 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
30000 |
0.17 |
| chr9_102403327_102403577 | 0.26 |
Gm22894 |
predicted gene, 22894 |
30334 |
0.14 |
| chr11_112374092_112374250 | 0.26 |
Gm11680 |
predicted gene 11680 |
266733 |
0.02 |
| chr7_30361694_30362080 | 0.26 |
Lrfn3 |
leucine rich repeat and fibronectin type III domain containing 3 |
885 |
0.31 |
| chr7_24595302_24595547 | 0.26 |
Zfp575 |
zinc finger protein 575 |
7783 |
0.08 |
| chr18_20114615_20114773 | 0.26 |
Dsc1 |
desmocollin 1 |
37 |
0.98 |
| chr1_46345449_46345605 | 0.26 |
Dnah7b |
dynein, axonemal, heavy chain 7B |
11093 |
0.21 |
| chr15_80680833_80681008 | 0.25 |
Fam83f |
family with sequence similarity 83, member F |
9073 |
0.13 |
| chr2_118554450_118554815 | 0.25 |
Bmf |
BCL2 modifying factor |
4945 |
0.18 |
| chr19_36675015_36675190 | 0.24 |
Hectd2os |
Hectd2, opposite strand |
14171 |
0.22 |
| chr3_133765600_133766533 | 0.24 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr4_105225226_105225391 | 0.24 |
Plpp3 |
phospholipid phosphatase 3 |
67961 |
0.13 |
| chr15_77831173_77831327 | 0.23 |
Gm22107 |
predicted gene, 22107 |
9506 |
0.15 |
| chr10_19062073_19062383 | 0.23 |
1700124M09Rik |
RIKEN cDNA 1700124M09 gene |
24950 |
0.14 |
| chr10_24087275_24087474 | 0.23 |
Taar8b |
trace amine-associated receptor 8B |
4920 |
0.1 |
| chr11_80429840_80430182 | 0.23 |
Psmd11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
124 |
0.96 |
| chr1_184873376_184873577 | 0.23 |
C130074G19Rik |
RIKEN cDNA C130074G19 gene |
9742 |
0.16 |
| chr16_91786325_91786478 | 0.23 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
2296 |
0.28 |
| chr2_103589515_103589686 | 0.23 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
23290 |
0.18 |
| chr6_90716699_90716865 | 0.23 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
253 |
0.91 |
| chr10_8086797_8087175 | 0.23 |
Gm48614 |
predicted gene, 48614 |
65694 |
0.11 |
| chr5_33934831_33935145 | 0.22 |
Nelfa |
negative elongation factor complex member A, Whsc2 |
1262 |
0.31 |
| chr6_72572233_72572384 | 0.22 |
Capg |
capping protein (actin filament), gelsolin-like |
16329 |
0.09 |
| chr6_135133072_135133223 | 0.22 |
Gm29803 |
predicted gene, 29803 |
1305 |
0.32 |
| chr2_164158645_164158808 | 0.22 |
n-R5s207 |
nuclear encoded rRNA 5S 207 |
1205 |
0.29 |
| chr2_151943059_151943241 | 0.22 |
Gm14154 |
predicted gene 14154 |
7491 |
0.14 |
| chr4_144913207_144913382 | 0.22 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
5585 |
0.23 |
| chr1_72182919_72183124 | 0.22 |
Mreg |
melanoregulin |
29286 |
0.12 |
| chr17_31303465_31303616 | 0.21 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
2003 |
0.24 |
| chr6_72604832_72604993 | 0.21 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
539 |
0.39 |
| chr9_48675944_48676095 | 0.21 |
Nnmt |
nicotinamide N-methyltransferase |
70866 |
0.11 |
| chr9_64795430_64795739 | 0.21 |
Dennd4a |
DENN/MADD domain containing 4A |
15756 |
0.19 |
| chr11_7203968_7204174 | 0.21 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
6289 |
0.18 |
| chr18_21220186_21220337 | 0.21 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
79862 |
0.08 |
| chr1_84059698_84059849 | 0.21 |
Pid1 |
phosphotyrosine interaction domain containing 1 |
6520 |
0.27 |
| chr8_35417462_35417628 | 0.21 |
Gm45301 |
predicted gene 45301 |
8039 |
0.17 |
| chr10_59466713_59466931 | 0.21 |
Mcu |
mitochondrial calcium uniporter |
8662 |
0.2 |
| chr15_74956771_74956940 | 0.20 |
Ly6e |
lymphocyte antigen 6 complex, locus E |
300 |
0.78 |
| chr10_61236047_61236201 | 0.20 |
Adamts14 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
37118 |
0.12 |
| chr6_54041419_54041593 | 0.20 |
Chn2 |
chimerin 2 |
1420 |
0.45 |
| chr11_101466397_101466548 | 0.20 |
Vat1 |
vesicle amine transport 1 |
242 |
0.75 |
| chr11_112810941_112811143 | 0.20 |
Gm11681 |
predicted gene 11681 |
12034 |
0.18 |
| chr3_91483300_91483476 | 0.20 |
S100a7l2 |
S100 calcium binding protein A7 like 2 |
392585 |
0.01 |
| chr11_86971141_86971293 | 0.20 |
Ypel2 |
yippee like 2 |
807 |
0.62 |
| chr4_144924031_144924237 | 0.20 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
5255 |
0.22 |
| chr11_101927534_101927685 | 0.19 |
Rpl27-ps2 |
ribosomal protein L27, pseudogene 2 |
11892 |
0.1 |
| chr4_144895225_144895561 | 0.19 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
2174 |
0.34 |
| chr1_74245666_74245817 | 0.19 |
Arpc2 |
actin related protein 2/3 complex, subunit 2 |
9191 |
0.09 |
| chr18_21373812_21373963 | 0.19 |
Gm22886 |
predicted gene, 22886 |
7044 |
0.2 |
| chr12_113221349_113221516 | 0.19 |
Gm25622 |
predicted gene, 25622 |
7283 |
0.14 |
| chr8_40996387_40996577 | 0.18 |
Mtus1 |
mitochondrial tumor suppressor 1 |
3723 |
0.2 |
| chr12_8004519_8005312 | 0.18 |
Apob |
apolipoprotein B |
7444 |
0.24 |
| chr14_22817406_22817579 | 0.18 |
Gm7473 |
predicted gene 7473 |
42248 |
0.2 |
| chr9_23030248_23030409 | 0.18 |
Bmper |
BMP-binding endothelial regulator |
192748 |
0.03 |
| chr13_17719442_17719753 | 0.18 |
Gm48621 |
predicted gene, 48621 |
9733 |
0.11 |
| chr19_30093012_30093163 | 0.18 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
1126 |
0.54 |
| chr6_5233850_5234020 | 0.18 |
Gm44250 |
predicted gene, 44250 |
13449 |
0.16 |
| chr2_103500304_103500503 | 0.18 |
Cat |
catalase |
15243 |
0.17 |
| chr16_86430347_86430530 | 0.18 |
Gm32357 |
predicted gene, 32357 |
194267 |
0.03 |
| chr12_40483542_40483866 | 0.18 |
Dock4 |
dedicator of cytokinesis 4 |
37368 |
0.17 |
| chr4_126091770_126091957 | 0.18 |
Oscp1 |
organic solute carrier partner 1 |
4184 |
0.13 |
| chr2_156175837_156176038 | 0.18 |
Rbm39 |
RNA binding motif protein 39 |
1446 |
0.28 |
| chr12_84203864_84204015 | 0.18 |
Gm31513 |
predicted gene, 31513 |
7970 |
0.11 |
| chr10_78324948_78325156 | 0.18 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
4362 |
0.1 |
| chr12_50524263_50524422 | 0.17 |
Prkd1 |
protein kinase D1 |
124756 |
0.06 |
| chr5_52982111_52982447 | 0.17 |
Gm30301 |
predicted gene, 30301 |
242 |
0.91 |
| chr5_28056784_28057184 | 0.17 |
Gm26608 |
predicted gene, 26608 |
1525 |
0.35 |
| chr8_120490974_120491151 | 0.17 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
2615 |
0.22 |
| chr4_117332720_117332922 | 0.17 |
Rnf220 |
ring finger protein 220 |
32793 |
0.11 |
| chr15_80645592_80645756 | 0.17 |
Mir6957 |
microRNA 6957 |
399 |
0.79 |
| chr2_24479227_24479538 | 0.17 |
Pax8 |
paired box 8 |
3783 |
0.16 |
| chr8_70719996_70720390 | 0.17 |
Gm3336 |
predicted gene 3336 |
1650 |
0.16 |
| chr6_105760602_105760775 | 0.17 |
Cntn4 |
contactin 4 |
82913 |
0.1 |
| chr4_65974196_65974358 | 0.17 |
Trim32 |
tripartite motif-containing 32 |
369028 |
0.01 |
| chr12_31944779_31944949 | 0.17 |
Hbp1 |
high mobility group box transcription factor 1 |
4457 |
0.22 |
| chr18_37953146_37953310 | 0.17 |
Hdac3 |
histone deacetylase 3 |
1690 |
0.17 |
| chr16_87512452_87512619 | 0.17 |
Gm24891 |
predicted gene, 24891 |
15630 |
0.12 |
| chr9_42293744_42293907 | 0.17 |
Gm36435 |
predicted gene, 36435 |
13947 |
0.15 |
| chr4_127077236_127077665 | 0.17 |
Zmym6 |
zinc finger, MYM-type 6 |
8 |
0.96 |
| chr17_29394463_29394614 | 0.17 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
18037 |
0.11 |
| chr12_108359338_108359496 | 0.17 |
Cyp46a1 |
cytochrome P450, family 46, subfamily a, polypeptide 1 |
7193 |
0.17 |
| chr15_36476020_36476181 | 0.16 |
Ankrd46 |
ankyrin repeat domain 46 |
20615 |
0.13 |
| chr4_58148098_58148270 | 0.16 |
Svep1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
58412 |
0.14 |
| chr8_127946458_127946625 | 0.16 |
Mir21c |
microRNA 21c |
331684 |
0.01 |
| chr1_160895233_160895410 | 0.16 |
Gm6177 |
predicted gene 6177 |
2201 |
0.15 |
| chr2_156311618_156311968 | 0.16 |
Cnbd2 |
cyclic nucleotide binding domain containing 2 |
506 |
0.53 |
| chr12_8003138_8004354 | 0.16 |
Apob |
apolipoprotein B |
8613 |
0.24 |
| chr19_3877298_3877449 | 0.16 |
Chka |
choline kinase alpha |
1968 |
0.16 |
| chr8_34941364_34941583 | 0.16 |
Tnks |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
22705 |
0.16 |
| chr5_130139764_130139934 | 0.16 |
Kctd7 |
potassium channel tetramerisation domain containing 7 |
5012 |
0.13 |
| chr11_22276015_22276166 | 0.16 |
Ehbp1 |
EH domain binding protein 1 |
9748 |
0.28 |
| chr5_8968601_8968752 | 0.16 |
Gm15610 |
predicted gene 15610 |
3272 |
0.16 |
| chr8_22404559_22404725 | 0.15 |
Gm45452 |
predicted gene 45452 |
1890 |
0.18 |
| chr19_46131357_46131554 | 0.15 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
442 |
0.73 |
| chr8_33801179_33801330 | 0.15 |
Rbpms |
RNA binding protein gene with multiple splicing |
4040 |
0.18 |
| chr7_78903289_78903499 | 0.15 |
Aen |
apoptosis enhancing nuclease |
815 |
0.5 |
| chr11_102208183_102208370 | 0.15 |
Hdac5 |
histone deacetylase 5 |
7460 |
0.09 |
| chr1_180177544_180177695 | 0.15 |
Coq8a |
coenzyme Q8A |
1012 |
0.47 |
| chr6_55001492_55001666 | 0.15 |
Ggct |
gamma-glutamyl cyclotransferase |
8629 |
0.17 |
| chr4_154159940_154160245 | 0.15 |
Tprgl |
transformation related protein 63 regulated like |
74 |
0.95 |
| chr1_182500136_182500458 | 0.15 |
Gm37069 |
predicted gene, 37069 |
6141 |
0.15 |
| chr13_93630762_93631092 | 0.15 |
Gm15622 |
predicted gene 15622 |
5545 |
0.17 |
| chr2_72907906_72908071 | 0.15 |
Sp3 |
trans-acting transcription factor 3 |
33096 |
0.14 |
| chr15_102171688_102171839 | 0.15 |
Csad |
cysteine sulfinic acid decarboxylase |
7256 |
0.1 |
| chr16_4701536_4701728 | 0.15 |
Nmral1 |
NmrA-like family domain containing 1 |
17456 |
0.1 |
| chr12_61927306_61927476 | 0.15 |
Gm24750 |
predicted gene, 24750 |
6817 |
0.22 |
| chr4_141751428_141751718 | 0.15 |
Agmat |
agmatine ureohydrolase (agmatinase) |
4901 |
0.14 |
| chr15_101191263_101191414 | 0.15 |
Mir6962 |
microRNA 6962 |
2531 |
0.16 |
| chr4_133130667_133130841 | 0.15 |
Wasf2 |
WAS protein family, member 2 |
125 |
0.96 |
| chr10_42987828_42987979 | 0.15 |
Gm47815 |
predicted gene, 47815 |
47090 |
0.12 |
| chr11_105281077_105281256 | 0.15 |
Mrc2 |
mannose receptor, C type 2 |
11477 |
0.19 |
| chrX_86250773_86250944 | 0.15 |
Mageb4 |
melanoma antigen, family B, 4 |
5361 |
0.18 |
| chr13_92740047_92740198 | 0.15 |
Gm5199 |
predicted gene 5199 |
20760 |
0.17 |
| chr5_114599320_114599476 | 0.15 |
Trpv4 |
transient receptor potential cation channel, subfamily V, member 4 |
31240 |
0.12 |
| chr3_96736661_96736837 | 0.15 |
Rnf115 |
ring finger protein 115 |
8910 |
0.08 |
| chr4_32507009_32507324 | 0.15 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
5661 |
0.25 |
| chr14_46061047_46061231 | 0.15 |
Gm37257 |
predicted gene, 37257 |
10 |
0.98 |
| chr15_66284513_66284672 | 0.15 |
Kcnq3 |
potassium voltage-gated channel, subfamily Q, member 3 |
1459 |
0.38 |
| chr6_37871246_37871403 | 0.15 |
Trim24 |
tripartite motif-containing 24 |
513 |
0.8 |
| chr5_122303802_122304107 | 0.15 |
Gm15842 |
predicted gene 15842 |
1532 |
0.26 |
| chr15_80081005_80081156 | 0.14 |
Rpl3 |
ribosomal protein L3 |
599 |
0.39 |
| chr3_157918436_157918587 | 0.14 |
Cth |
cystathionase (cystathionine gamma-lyase) |
6540 |
0.14 |
| chr19_4878250_4878444 | 0.14 |
Zdhhc24 |
zinc finger, DHHC domain containing 24 |
321 |
0.53 |
| chr4_61590678_61590958 | 0.14 |
Mup17 |
major urinary protein 17 |
5053 |
0.19 |
| chr7_65370516_65370694 | 0.14 |
Tjp1 |
tight junction protein 1 |
410 |
0.84 |
| chr1_36546473_36546624 | 0.14 |
Ankrd39 |
ankyrin repeat domain 39 |
671 |
0.47 |
| chr15_75994590_75994817 | 0.14 |
Mapk15 |
mitogen-activated protein kinase 15 |
934 |
0.32 |
| chr14_118124640_118124836 | 0.14 |
Tgds |
TDP-glucose 4,6-dehydratase |
8006 |
0.18 |
| chr11_86952607_86952758 | 0.14 |
Ypel2 |
yippee like 2 |
19342 |
0.18 |
| chr6_121561006_121561170 | 0.14 |
Gm43934 |
predicted gene, 43934 |
887 |
0.55 |
| chr6_51270618_51270787 | 0.14 |
Mir148a |
microRNA 148a |
792 |
0.68 |
| chr10_74935143_74935297 | 0.14 |
Gnaz |
guanine nucleotide binding protein, alpha z subunit |
31957 |
0.17 |
| chr13_34725685_34725960 | 0.14 |
Gm47151 |
predicted gene, 47151 |
5071 |
0.13 |
| chr17_31788157_31788318 | 0.14 |
Gm49999 |
predicted gene, 49999 |
1267 |
0.43 |
| chr11_18900383_18900558 | 0.14 |
2900018N21Rik |
RIKEN cDNA 2900018N21 gene |
10630 |
0.16 |
| chr19_32534401_32535184 | 0.14 |
Gm36419 |
predicted gene, 36419 |
8556 |
0.18 |
| chr11_72281229_72281388 | 0.14 |
n-R5s70 |
nuclear encoded rRNA 5S 70 |
292 |
0.83 |
| chr18_36713491_36713783 | 0.14 |
Cd14 |
CD14 antigen |
13101 |
0.07 |
| chr6_124453188_124453388 | 0.14 |
Clstn3 |
calsyntenin 3 |
3116 |
0.14 |
| chr9_59470341_59470523 | 0.14 |
Rps11-ps1 |
ribosomal protein S11, pseudogene 1 |
1801 |
0.31 |
| chr1_72837567_72837745 | 0.14 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
11043 |
0.21 |
| chr2_164562758_164562924 | 0.14 |
Wfdc2 |
WAP four-disulfide core domain 2 |
127 |
0.9 |
| chr8_120619240_120619416 | 0.14 |
1190005I06Rik |
RIKEN cDNA 1190005I06 gene |
15035 |
0.09 |
| chr13_98693378_98693698 | 0.14 |
Tmem171 |
transmembrane protein 171 |
1230 |
0.37 |
| chr17_5840748_5840949 | 0.14 |
Snx9 |
sorting nexin 9 |
481 |
0.74 |
| chr12_81595637_81595788 | 0.13 |
Med6 |
mediator complex subunit 6 |
704 |
0.53 |
| chr5_64542110_64542274 | 0.13 |
Gm43836 |
predicted gene 43836 |
15991 |
0.12 |
| chr12_83474032_83474213 | 0.13 |
Dpf3 |
D4, zinc and double PHD fingers, family 3 |
13586 |
0.18 |
| chr4_55242971_55243122 | 0.13 |
Gm12508 |
predicted gene 12508 |
11694 |
0.17 |
| chr4_119117332_119117542 | 0.13 |
Slc2a1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
8523 |
0.1 |
| chr12_111894295_111894473 | 0.13 |
Ppp1r13b |
protein phosphatase 1, regulatory subunit 13B |
13235 |
0.12 |
| chr6_17319461_17319624 | 0.13 |
Cav1 |
caveolin 1, caveolae protein |
11819 |
0.18 |
| chr11_97958258_97958415 | 0.13 |
Gm11633 |
predicted gene 11633 |
10456 |
0.1 |
| chr12_104355745_104355928 | 0.13 |
Serpina3l-ps |
serine (or cysteine) peptidase inhibitor, clade A, member 3L, pseudogene |
2167 |
0.21 |
| chr1_74190938_74191089 | 0.13 |
Cxcr1 |
chemokine (C-X-C motif) receptor 1 |
2222 |
0.19 |
| chr4_11089211_11089550 | 0.13 |
Ndufaf6 |
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
13175 |
0.14 |
| chr13_42341987_42342149 | 0.13 |
Gm47118 |
predicted gene, 47118 |
37077 |
0.17 |
| chr7_28801073_28801224 | 0.13 |
Rinl |
Ras and Rab interactor-like |
3250 |
0.1 |
| chr5_134853069_134853266 | 0.13 |
Tmem270 |
transmembrane protein 270 |
53566 |
0.07 |
| chr12_104798010_104798169 | 0.13 |
Clmn |
calmin |
17218 |
0.19 |
| chr1_105891390_105891564 | 0.13 |
Gm37779 |
predicted gene, 37779 |
465 |
0.82 |
| chr4_120666699_120666867 | 0.13 |
Cited4 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
211 |
0.92 |
| chr5_123791025_123791189 | 0.13 |
Kntc1 |
kinetochore associated 1 |
3895 |
0.18 |
| chr5_76306348_76306506 | 0.13 |
Gm7467 |
predicted gene 7467 |
1551 |
0.23 |
| chr4_60416791_60417110 | 0.13 |
Mup9 |
major urinary protein 9 |
3634 |
0.18 |
| chr1_111864068_111864581 | 0.13 |
Dsel |
dermatan sulfate epimerase-like |
444 |
0.83 |
| chr12_100989931_100990090 | 0.13 |
Gm36756 |
predicted gene, 36756 |
2962 |
0.17 |
| chr17_63497841_63498445 | 0.13 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
1874 |
0.43 |
| chr8_107483340_107483510 | 0.13 |
Wwp2 |
WW domain containing E3 ubiquitin protein ligase 2 |
93 |
0.97 |
| chr5_37874933_37875104 | 0.13 |
Gm20052 |
predicted gene, 20052 |
18754 |
0.19 |
| chr7_141569233_141569384 | 0.13 |
Ap2a2 |
adaptor-related protein complex 2, alpha 2 subunit |
6778 |
0.12 |
| chr14_114418353_114418542 | 0.13 |
Gm19829 |
predicted gene, 19829 |
107644 |
0.08 |
| chr13_48956580_48956936 | 0.13 |
Fam120a |
family with sequence similarity 120, member A |
11259 |
0.23 |
| chr16_22958475_22958991 | 0.13 |
Hrg |
histidine-rich glycoprotein |
7633 |
0.12 |
| chr11_72243451_72243602 | 0.13 |
1700051A21Rik |
RIKEN cDNA 1700051A21 gene |
22895 |
0.09 |
| chr14_68361550_68361701 | 0.13 |
Gm31227 |
predicted gene, 31227 |
100006 |
0.07 |
| chr9_57758758_57758995 | 0.13 |
Clk3 |
CDC-like kinase 3 |
3555 |
0.17 |
| chr8_115757553_115757722 | 0.13 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
49843 |
0.16 |
| chr3_142191728_142191879 | 0.13 |
Gm6059 |
predicted gene 6059 |
7052 |
0.23 |
| chr17_26610962_26611350 | 0.13 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
2294 |
0.2 |
| chr6_54850083_54850243 | 0.12 |
Znrf2 |
zinc and ring finger 2 |
32715 |
0.16 |
| chr9_37211963_37212270 | 0.12 |
Tmem218 |
transmembrane protein 218 |
3893 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.2 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0006057 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.0 | 0.0 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0018637 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0003933 | GTP cyclohydrolase activity(GO:0003933) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |