Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
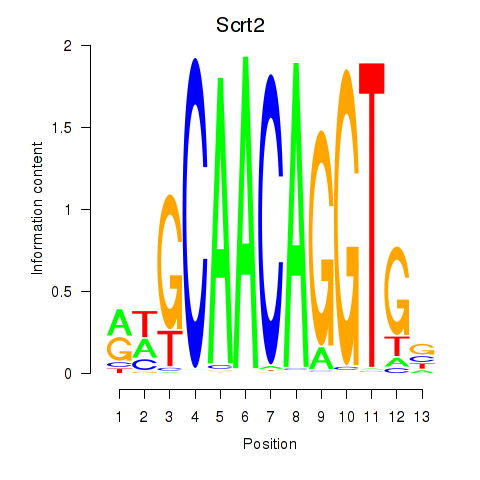
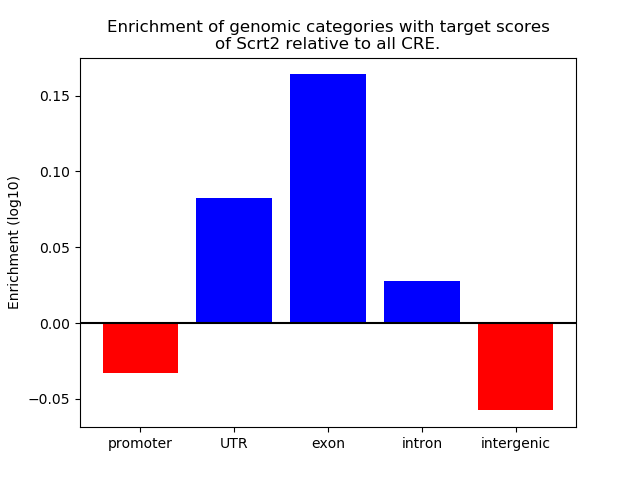
Results for Scrt2
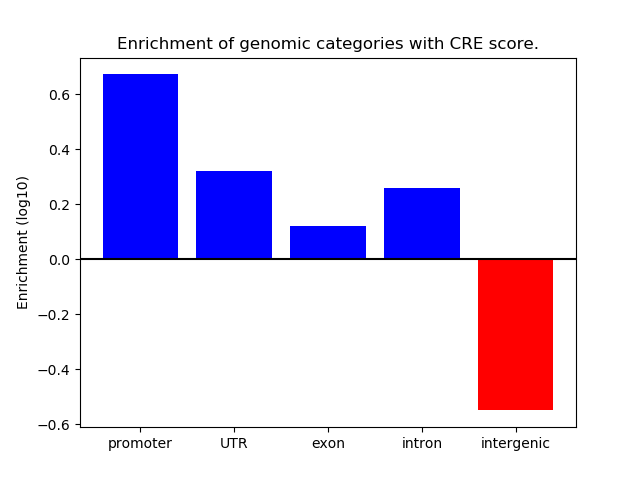
Z-value: 0.62
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSMUSG00000060257.2 | scratch family zinc finger 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_152089191_152089351 | Scrt2 | 7742 | 0.140363 | -0.05 | 9.2e-01 | Click! |
| chr2_152081883_152082034 | Scrt2 | 429 | 0.776922 | -0.05 | 9.2e-01 | Click! |
Activity of the Scrt2 motif across conditions
Conditions sorted by the z-value of the Scrt2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
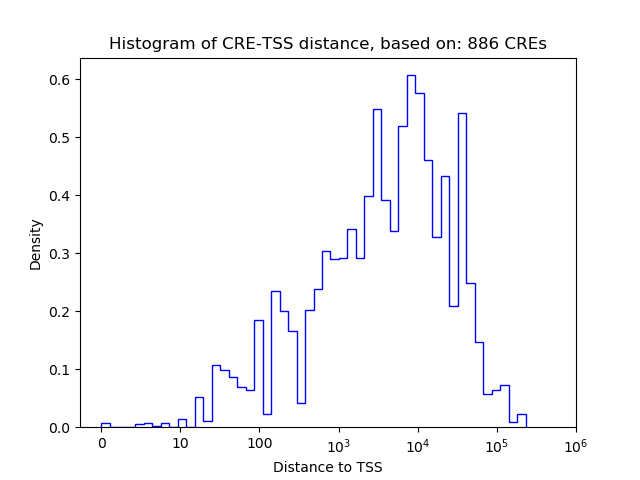
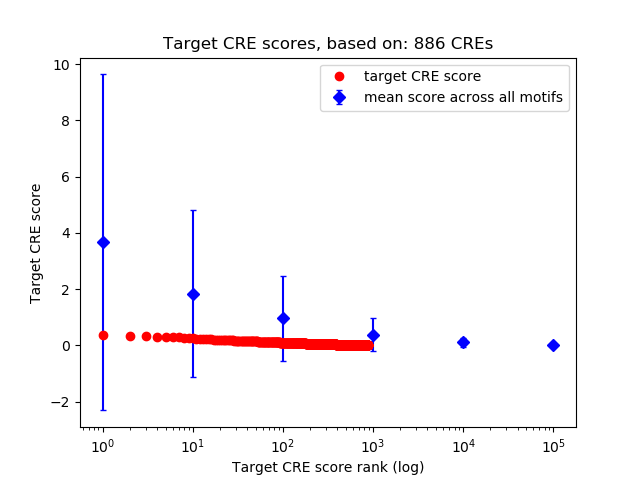
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_5091846_5091997 | 0.35 |
Svil |
supervillin |
2460 |
0.4 |
| chr2_148014305_148014474 | 0.32 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
23881 |
0.16 |
| chr11_103230027_103230201 | 0.32 |
Map3k14 |
mitogen-activated protein kinase kinase kinase 14 |
8267 |
0.12 |
| chr3_94832747_94832920 | 0.29 |
Gm15263 |
predicted gene 15263 |
144 |
0.93 |
| chr8_86807888_86808046 | 0.29 |
N4bp1 |
NEDD4 binding protein 1 |
36078 |
0.13 |
| chr2_42022841_42022992 | 0.28 |
Gm13461 |
predicted gene 13461 |
34087 |
0.23 |
| chr5_8189740_8189891 | 0.28 |
Gm21759 |
predicted gene, 21759 |
10179 |
0.18 |
| chr11_5326373_5326703 | 0.26 |
Gm30172 |
predicted gene, 30172 |
52453 |
0.1 |
| chr9_66200038_66200209 | 0.25 |
Dapk2 |
death-associated protein kinase 2 |
20624 |
0.19 |
| chr3_84657319_84657504 | 0.24 |
Tmem154 |
transmembrane protein 154 |
8781 |
0.23 |
| chr8_95800469_95800620 | 0.24 |
4930513N10Rik |
RIKEN cDNA 4930513N10 gene |
6200 |
0.09 |
| chr7_118698593_118698744 | 0.22 |
Gde1 |
glycerophosphodiester phosphodiesterase 1 |
6712 |
0.13 |
| chr11_120658973_120659131 | 0.22 |
Notumos |
notum palmitoleoyl-protein carboxylesterase, opposite strand |
105 |
0.86 |
| chr7_14428951_14429109 | 0.22 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
7417 |
0.17 |
| chr15_57693174_57693501 | 0.22 |
Zhx2 |
zinc fingers and homeoboxes 2 |
1328 |
0.47 |
| chr1_164415295_164415654 | 0.21 |
Gm37411 |
predicted gene, 37411 |
9239 |
0.16 |
| chr3_89101600_89101915 | 0.21 |
Fdps |
farnesyl diphosphate synthetase |
95 |
0.86 |
| chr10_33789536_33789687 | 0.20 |
Sult3a2 |
sulfotransferase family 3A, member 2 |
2907 |
0.22 |
| chr9_107588315_107588481 | 0.20 |
Ifrd2 |
interferon-related developmental regulator 2 |
641 |
0.32 |
| chr6_136522266_136522417 | 0.20 |
Gm44140 |
predicted gene, 44140 |
3310 |
0.17 |
| chr7_45711383_45711838 | 0.20 |
Sphk2 |
sphingosine kinase 2 |
1833 |
0.12 |
| chr19_10596220_10596418 | 0.19 |
Tkfc |
triokinase, FMN cyclase |
7939 |
0.09 |
| chr16_43396888_43397060 | 0.19 |
Gm15713 |
predicted gene 15713 |
23222 |
0.16 |
| chrX_102187134_102187366 | 0.19 |
Rps4x |
ribosomal protein S4, X-linked |
513 |
0.68 |
| chr19_32249385_32249564 | 0.19 |
Sgms1 |
sphingomyelin synthase 1 |
1465 |
0.48 |
| chr11_94281352_94281503 | 0.18 |
Gm21885 |
predicted gene, 21885 |
446 |
0.78 |
| chr4_57070625_57070924 | 0.18 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
215 |
0.95 |
| chr5_145918586_145918760 | 0.18 |
Gm42431 |
predicted gene 42431 |
205 |
0.92 |
| chr2_121173551_121173702 | 0.17 |
Tubgcp4 |
tubulin, gamma complex associated protein 4 |
25 |
0.96 |
| chr4_150559587_150559790 | 0.17 |
Gm13091 |
predicted gene 13091 |
8856 |
0.21 |
| chr5_151223190_151223357 | 0.17 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
10501 |
0.21 |
| chr11_30773928_30774079 | 0.17 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
1682 |
0.31 |
| chr7_67634879_67635046 | 0.17 |
Gm45100 |
predicted gene 45100 |
2803 |
0.19 |
| chr1_134564053_134564250 | 0.16 |
Kdm5b |
lysine (K)-specific demethylase 5B |
3944 |
0.16 |
| chr18_20661522_20661684 | 0.16 |
Gm16090 |
predicted gene 16090 |
3657 |
0.2 |
| chr3_153198701_153198852 | 0.16 |
1700094M23Rik |
RIKEN cDNA 1700094M23 gene |
15961 |
0.2 |
| chr11_5444005_5444193 | 0.16 |
Znrf3 |
zinc and ring finger 3 |
748 |
0.65 |
| chr4_63553855_63554016 | 0.16 |
Tmem268 |
transmembrane protein 268 |
4846 |
0.13 |
| chr3_95801499_95801680 | 0.15 |
Gm17815 |
predicted gene, 17815 |
3266 |
0.11 |
| chr11_4187555_4187772 | 0.15 |
Tbc1d10a |
TBC1 domain family, member 10a |
841 |
0.42 |
| chr8_77024149_77024322 | 0.15 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
103778 |
0.07 |
| chr6_134791299_134791459 | 0.15 |
Dusp16 |
dual specificity phosphatase 16 |
45 |
0.97 |
| chr15_68205229_68205387 | 0.15 |
Zfat |
zinc finger and AT hook domain containing |
53479 |
0.13 |
| chr1_178600841_178601326 | 0.15 |
Gm24405 |
predicted gene, 24405 |
35822 |
0.18 |
| chr2_146577293_146577459 | 0.14 |
4933406D12Rik |
RIKEN cDNA 4933406D12 gene |
34445 |
0.2 |
| chr14_16573464_16573628 | 0.14 |
Rarb |
retinoic acid receptor, beta |
1499 |
0.4 |
| chr11_42999314_42999478 | 0.14 |
Gm9972 |
predicted gene 9972 |
36350 |
0.21 |
| chr10_40164480_40164639 | 0.14 |
Gm31992 |
predicted gene, 31992 |
449 |
0.75 |
| chr12_57650105_57650438 | 0.14 |
Ttc6 |
tetratricopeptide repeat domain 6 |
856 |
0.55 |
| chr8_47989535_47989869 | 0.14 |
Wwc2 |
WW, C2 and coiled-coil domain containing 2 |
849 |
0.62 |
| chr8_115832098_115832272 | 0.14 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
124391 |
0.06 |
| chr8_41057877_41058089 | 0.14 |
Mtus1 |
mitochondrial tumor suppressor 1 |
3189 |
0.2 |
| chr4_155566693_155567032 | 0.13 |
Nadk |
NAD kinase |
2790 |
0.15 |
| chr2_147321117_147321284 | 0.13 |
Gm14110 |
predicted gene 14110 |
3887 |
0.19 |
| chr5_124172367_124172532 | 0.13 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
12669 |
0.12 |
| chr5_125492454_125492640 | 0.13 |
Gm27551 |
predicted gene, 27551 |
13170 |
0.13 |
| chr18_20114615_20114773 | 0.13 |
Dsc1 |
desmocollin 1 |
37 |
0.98 |
| chr2_59158255_59158412 | 0.13 |
Ccdc148 |
coiled-coil domain containing 148 |
2201 |
0.27 |
| chr10_57285621_57285792 | 0.12 |
Gm7001 |
predicted gene 7001 |
5183 |
0.23 |
| chr2_156091789_156091940 | 0.12 |
Gm28036 |
predicted gene, 28036 |
5064 |
0.1 |
| chr17_72795441_72795610 | 0.12 |
Ypel5 |
yippee like 5 |
40928 |
0.18 |
| chr5_142421964_142422141 | 0.12 |
Foxk1 |
forkhead box K1 |
20552 |
0.19 |
| chr10_86339756_86339912 | 0.12 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
36980 |
0.16 |
| chr12_40037314_40037592 | 0.12 |
Arl4a |
ADP-ribosylation factor-like 4A |
24 |
0.93 |
| chr6_94225898_94226102 | 0.12 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
57025 |
0.14 |
| chr19_10193896_10194210 | 0.12 |
Fads1 |
fatty acid desaturase 1 |
734 |
0.46 |
| chr5_24826145_24826327 | 0.12 |
Rheb |
Ras homolog enriched in brain |
12277 |
0.16 |
| chr12_108359338_108359496 | 0.12 |
Cyp46a1 |
cytochrome P450, family 46, subfamily a, polypeptide 1 |
7193 |
0.17 |
| chr8_84147166_84147595 | 0.12 |
Cc2d1a |
coiled-coil and C2 domain containing 1A |
485 |
0.42 |
| chr5_88645260_88645648 | 0.12 |
Rufy3 |
RUN and FYVE domain containing 3 |
1237 |
0.44 |
| chr10_68096521_68096741 | 0.12 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
39995 |
0.15 |
| chr9_44485795_44485985 | 0.12 |
Bcl9l |
B cell CLL/lymphoma 9-like |
1357 |
0.17 |
| chr2_121419496_121419659 | 0.11 |
Pdia3 |
protein disulfide isomerase associated 3 |
5053 |
0.08 |
| chr15_82703396_82703582 | 0.11 |
Cyp2d38-ps |
cytochrome P450, family 2, subfamily d, member 38, pseudogene |
3776 |
0.11 |
| chrX_77511793_77512164 | 0.11 |
Tbl1x |
transducin (beta)-like 1 X-linked |
764 |
0.71 |
| chr12_106326776_106326927 | 0.11 |
Gm17032 |
predicted gene 17032 |
517 |
0.85 |
| chr6_54680979_54681130 | 0.11 |
Mturn |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
570 |
0.56 |
| chr3_108314174_108314346 | 0.11 |
Gm43099 |
predicted gene 43099 |
26379 |
0.07 |
| chr13_31561603_31562246 | 0.11 |
A530084C06Rik |
RIKEN cDNA A530084C06 gene |
2591 |
0.18 |
| chrX_139665962_139666117 | 0.11 |
Gm6322 |
predicted gene 6322 |
11450 |
0.17 |
| chr13_37913218_37913408 | 0.11 |
Rreb1 |
ras responsive element binding protein 1 |
24442 |
0.18 |
| chr15_25663250_25663426 | 0.11 |
Myo10 |
myosin X |
11172 |
0.18 |
| chr15_96371640_96371833 | 0.11 |
Arid2 |
AT rich interactive domain 2 (ARID, RFX-like) |
7078 |
0.21 |
| chrX_140567606_140567780 | 0.11 |
AL683809.1 |
TSC22 domain family, member 3 (Tsc22d3), pseuodgene |
15906 |
0.17 |
| chr12_71925163_71925487 | 0.11 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
35595 |
0.17 |
| chr15_38328239_38328390 | 0.11 |
Gm35248 |
predicted gene, 35248 |
16021 |
0.14 |
| chr1_91445260_91445618 | 0.10 |
Per2 |
period circadian clock 2 |
4426 |
0.13 |
| chr2_37578235_37578422 | 0.10 |
Strbp |
spermatid perinuclear RNA binding protein |
8190 |
0.18 |
| chr8_85493343_85493503 | 0.10 |
Gpt2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
814 |
0.6 |
| chr6_54151964_54152130 | 0.10 |
9530036M11Rik |
RIKEN cDNA 9530036M11 gene |
1110 |
0.53 |
| chr7_15947981_15948207 | 0.10 |
Nop53 |
NOP53 ribosome biogenesis factor |
2020 |
0.18 |
| chr19_53225616_53225797 | 0.10 |
Add3 |
adducin 3 (gamma) |
27660 |
0.12 |
| chr11_72243451_72243602 | 0.10 |
1700051A21Rik |
RIKEN cDNA 1700051A21 gene |
22895 |
0.09 |
| chr11_60422153_60422315 | 0.10 |
Atpaf2 |
ATP synthase mitochondrial F1 complex assembly factor 2 |
3777 |
0.13 |
| chr6_149142584_149142797 | 0.10 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
1044 |
0.42 |
| chr8_3262136_3262316 | 0.10 |
Gm16180 |
predicted gene 16180 |
2522 |
0.28 |
| chr5_66069722_66069897 | 0.10 |
Gm43775 |
predicted gene 43775 |
3086 |
0.17 |
| chr2_144265873_144266024 | 0.10 |
Snord17 |
small nucleolar RNA, C/D box 17 |
268 |
0.82 |
| chr7_99634093_99634268 | 0.10 |
Tpbgl |
trophoblast glycoprotein-like |
7077 |
0.1 |
| chr2_9889364_9889515 | 0.10 |
9230102O04Rik |
RIKEN cDNA 9230102O04 gene |
101 |
0.87 |
| chr12_84399769_84399951 | 0.10 |
Entpd5 |
ectonucleoside triphosphate diphosphohydrolase 5 |
1025 |
0.38 |
| chr2_6352628_6352806 | 0.10 |
Usp6nl |
USP6 N-terminal like |
16 |
0.98 |
| chr5_74833941_74834092 | 0.10 |
Gm17906 |
predicted gene, 17906 |
49319 |
0.12 |
| chr10_84757574_84757782 | 0.09 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
1616 |
0.43 |
| chr4_41707202_41707379 | 0.09 |
Rpp25l |
ribonuclease P/MRP 25 subunit-like |
6244 |
0.09 |
| chr13_46930435_46930600 | 0.09 |
Kif13a |
kinesin family member 13A |
650 |
0.61 |
| chr11_87697419_87697735 | 0.09 |
Rnf43 |
ring finger protein 43 |
2320 |
0.16 |
| chr6_33060273_33060430 | 0.09 |
Chchd3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
91 |
0.97 |
| chr3_84459628_84459793 | 0.09 |
Fhdc1 |
FH2 domain containing 1 |
5947 |
0.25 |
| chr19_7382057_7382208 | 0.09 |
1700105P06Rik |
RIKEN cDNA 1700105P06 gene |
405 |
0.6 |
| chr18_56431981_56432145 | 0.09 |
Gramd3 |
GRAM domain containing 3 |
69 |
0.98 |
| chr15_78830923_78831096 | 0.09 |
Gm7318 |
predicted gene 7318 |
10199 |
0.09 |
| chr5_33337165_33337512 | 0.09 |
Maea |
macrophage erythroblast attacher |
1153 |
0.44 |
| chr9_106231818_106232229 | 0.09 |
Alas1 |
aminolevulinic acid synthase 1 |
5061 |
0.11 |
| chr1_193928644_193928965 | 0.09 |
Gm21362 |
predicted gene, 21362 |
61795 |
0.15 |
| chr2_160990763_160990931 | 0.09 |
Chd6 |
chromodomain helicase DNA binding protein 6 |
5801 |
0.18 |
| chr2_104849084_104849595 | 0.09 |
Prrg4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
154 |
0.94 |
| chr2_121442121_121442548 | 0.09 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
267 |
0.78 |
| chr14_69325074_69325267 | 0.09 |
Gm16677 |
predicted gene, 16677 |
11912 |
0.09 |
| chr10_67288327_67288543 | 0.09 |
Nrbf2 |
nuclear receptor binding factor 2 |
3130 |
0.22 |
| chr14_69543328_69543511 | 0.09 |
Gm27174 |
predicted gene 27174 |
11913 |
0.1 |
| chr7_144550697_144551074 | 0.09 |
Ppfia1 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
1477 |
0.36 |
| chr4_108074336_108074558 | 0.08 |
Scp2 |
sterol carrier protein 2, liver |
3028 |
0.19 |
| chr5_96983306_96983462 | 0.08 |
Gm9484 |
predicted gene 9484 |
13980 |
0.12 |
| chr3_129901851_129902495 | 0.08 |
Casp6 |
caspase 6 |
672 |
0.65 |
| chr7_52006040_52006551 | 0.08 |
Svip |
small VCP/p97-interacting protein |
277 |
0.9 |
| chr1_190822749_190822917 | 0.08 |
Rps6kc1 |
ribosomal protein S6 kinase polypeptide 1 |
11203 |
0.24 |
| chr9_43594764_43595309 | 0.08 |
Gm29909 |
predicted gene, 29909 |
13148 |
0.18 |
| chr8_71521779_71521941 | 0.08 |
Gm7850 |
predicted gene 7850 |
1714 |
0.15 |
| chr18_53864446_53864629 | 0.08 |
Csnk1g3 |
casein kinase 1, gamma 3 |
2349 |
0.42 |
| chr12_8003138_8004354 | 0.08 |
Apob |
apolipoprotein B |
8613 |
0.24 |
| chr6_138075354_138075525 | 0.08 |
Slc15a5 |
solute carrier family 15, member 5 |
2243 |
0.41 |
| chr9_78107923_78108247 | 0.08 |
Fbxo9 |
f-box protein 9 |
502 |
0.62 |
| chr10_68180038_68180189 | 0.08 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
43487 |
0.17 |
| chrX_142409444_142409630 | 0.08 |
Gm6067 |
predicted gene 6067 |
3096 |
0.18 |
| chr12_71875530_71876050 | 0.08 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
13940 |
0.21 |
| chr3_68493220_68493423 | 0.08 |
Schip1 |
schwannomin interacting protein 1 |
170 |
0.96 |
| chr7_26087500_26087684 | 0.08 |
Gm29920 |
predicted gene, 29920 |
12724 |
0.13 |
| chr8_95716082_95716671 | 0.08 |
Setd6 |
SET domain containing 6 |
214 |
0.87 |
| chr7_143501066_143501235 | 0.08 |
Phlda2 |
pleckstrin homology like domain, family A, member 2 |
1391 |
0.29 |
| chr13_63365393_63365551 | 0.08 |
Fancc |
Fanconi anemia, complementation group C |
13300 |
0.11 |
| chr15_38422882_38423097 | 0.08 |
Gm41307 |
predicted gene, 41307 |
18835 |
0.15 |
| chr2_26560809_26560960 | 0.08 |
Gm13358 |
predicted gene 13358 |
2259 |
0.16 |
| chr11_5346895_5347220 | 0.08 |
Znrf3 |
zinc and ring finger 3 |
34683 |
0.15 |
| chr3_122984334_122984522 | 0.08 |
Usp53 |
ubiquitin specific peptidase 53 |
43 |
0.96 |
| chr16_56715953_56716104 | 0.08 |
Tfg |
Trk-fused gene |
1154 |
0.55 |
| chr15_103837588_103837739 | 0.08 |
Gm21178 |
predicted gene, 21178 |
10411 |
0.17 |
| chrX_160414816_160415110 | 0.08 |
Adgrg2 |
adhesion G protein-coupled receptor G2 |
12329 |
0.21 |
| chr15_7172806_7173149 | 0.08 |
Lifr |
LIF receptor alpha |
18624 |
0.24 |
| chr7_140971847_140972117 | 0.07 |
Ifitm1 |
interferon induced transmembrane protein 1 |
3942 |
0.09 |
| chr11_64188199_64188501 | 0.07 |
Gm12290 |
predicted gene 12290 |
23777 |
0.24 |
| chr16_76354348_76354578 | 0.07 |
Nrip1 |
nuclear receptor interacting protein 1 |
18574 |
0.19 |
| chr19_24999693_24999865 | 0.07 |
Dock8 |
dedicator of cytokinesis 8 |
235 |
0.93 |
| chr13_3768673_3768881 | 0.07 |
Gm35190 |
predicted gene, 35190 |
2055 |
0.25 |
| chr11_104526046_104526207 | 0.07 |
Cdc27 |
cell division cycle 27 |
12850 |
0.11 |
| chr5_82521778_82521958 | 0.07 |
Rpl7-ps7 |
ribosomal protein L7, pseudogene 7 |
185017 |
0.03 |
| chr18_50717950_50718102 | 0.07 |
Pudp |
pseudouridine 5'-phosphatase |
116607 |
0.07 |
| chr11_80168670_80168836 | 0.07 |
Adap2os |
ArfGAP with dual PH domains 2, opposite strand |
8876 |
0.13 |
| chr2_27595395_27595595 | 0.07 |
Gm13421 |
predicted gene 13421 |
55066 |
0.1 |
| chr8_83585276_83585468 | 0.07 |
Gm45823 |
predicted gene 45823 |
153 |
0.9 |
| chr17_12927243_12927577 | 0.07 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
272 |
0.78 |
| chr3_50279626_50279824 | 0.07 |
Gm2345 |
predicted gene 2345 |
24870 |
0.22 |
| chr4_121023495_121023646 | 0.07 |
Smap2 |
small ArfGAP 2 |
6323 |
0.13 |
| chr1_165461123_165461290 | 0.07 |
Mpc2 |
mitochondrial pyruvate carrier 2 |
18 |
0.92 |
| chr19_34877607_34877783 | 0.07 |
Pank1 |
pantothenate kinase 1 |
154 |
0.95 |
| chr6_54388576_54388751 | 0.07 |
9130019P16Rik |
RIKEN cDNA 9130019P16 gene |
40874 |
0.13 |
| chr4_11447041_11447202 | 0.07 |
Gm11822 |
predicted gene 11822 |
26357 |
0.13 |
| chr13_101523481_101523756 | 0.07 |
Gm47533 |
predicted gene, 47533 |
21588 |
0.16 |
| chr5_33629008_33629185 | 0.07 |
Fam53a |
family with sequence similarity 53, member A |
34 |
0.93 |
| chr7_27032583_27032938 | 0.07 |
Cyp2a12 |
cytochrome P450, family 2, subfamily a, polypeptide 12 |
3670 |
0.15 |
| chr7_25987036_25987409 | 0.07 |
Gm45225 |
predicted gene 45225 |
5720 |
0.14 |
| chr11_58982009_58982210 | 0.07 |
Trim11 |
tripartite motif-containing 11 |
3993 |
0.08 |
| chr19_37441259_37441628 | 0.07 |
Hhex |
hematopoietically expressed homeobox |
4704 |
0.13 |
| chr8_85432635_85432800 | 0.07 |
1700051O22Rik |
RIKEN cDNA 1700051O22 Gene |
31 |
0.57 |
| chr11_100398084_100398235 | 0.07 |
Jup |
junction plakoglobin |
396 |
0.67 |
| chr1_92083274_92083514 | 0.07 |
Hdac4 |
histone deacetylase 4 |
29937 |
0.19 |
| chr7_100658121_100658314 | 0.07 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
147 |
0.93 |
| chr10_87552519_87552755 | 0.06 |
Pah |
phenylalanine hydroxylase |
5964 |
0.24 |
| chr17_72957167_72957340 | 0.06 |
Gm23649 |
predicted gene, 23649 |
30545 |
0.17 |
| chrX_152769919_152770108 | 0.06 |
Shroom2 |
shroom family member 2 |
548 |
0.79 |
| chr7_143190080_143190255 | 0.06 |
Gm28821 |
predicted gene 28821 |
49559 |
0.09 |
| chr5_108038198_108038358 | 0.06 |
Rps19-ps8 |
ribosomal protein S19, pseudogene 8 |
17638 |
0.13 |
| chr19_38077879_38078207 | 0.06 |
Gm23300 |
predicted gene, 23300 |
6114 |
0.14 |
| chr7_73372055_73372220 | 0.06 |
Rgma |
repulsive guidance molecule family member A |
3372 |
0.17 |
| chrX_103994856_103995029 | 0.06 |
Gm14892 |
predicted gene 14892 |
6395 |
0.15 |
| chr6_108314149_108314309 | 0.06 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
42025 |
0.18 |
| chr10_96742974_96743161 | 0.06 |
Gm6859 |
predicted gene 6859 |
26761 |
0.15 |
| chr15_12824672_12824833 | 0.06 |
Drosha |
drosha, ribonuclease type III |
63 |
0.55 |
| chr1_88272907_88273179 | 0.06 |
Hjurp |
Holliday junction recognition protein |
76 |
0.95 |
| chr2_170459898_170460068 | 0.06 |
Gm14269 |
predicted gene 14269 |
3836 |
0.19 |
| chr18_35033854_35034028 | 0.06 |
Gm36037 |
predicted gene, 36037 |
50971 |
0.09 |
| chr4_105183113_105183302 | 0.06 |
Plpp3 |
phospholipid phosphatase 3 |
25860 |
0.23 |
| chr14_16364204_16364368 | 0.06 |
Top2b |
topoisomerase (DNA) II beta |
893 |
0.52 |
| chr3_135290385_135290563 | 0.06 |
Bdh2 |
3-hydroxybutyrate dehydrogenase, type 2 |
1816 |
0.29 |
| chr7_145104659_145104815 | 0.06 |
Gm45181 |
predicted gene 45181 |
58259 |
0.12 |
| chr17_15381768_15382106 | 0.06 |
Dll1 |
delta like canonical Notch ligand 1 |
5065 |
0.17 |
| chr1_45876552_45876725 | 0.06 |
Gm5269 |
predicted gene 5269 |
13440 |
0.12 |
| chr14_55721204_55721397 | 0.06 |
Rabggta |
Rab geranylgeranyl transferase, a subunit |
876 |
0.3 |
| chr6_71166909_71167070 | 0.06 |
Gm44427 |
predicted gene, 44427 |
2190 |
0.22 |
| chr2_4586318_4586682 | 0.06 |
Gm13179 |
predicted gene 13179 |
787 |
0.66 |
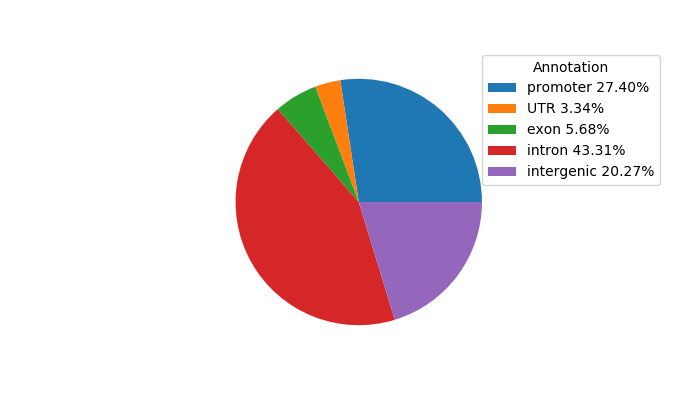
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004337 | geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |