Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
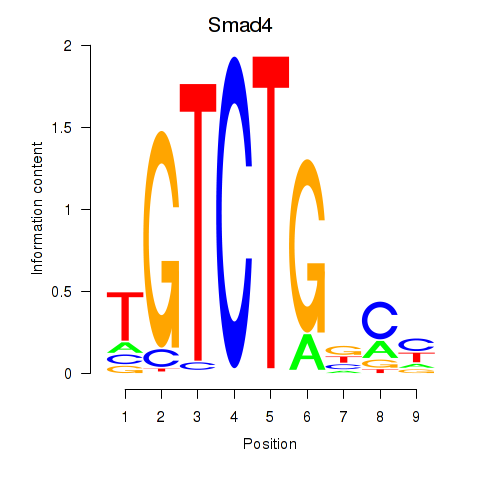
Results for Smad4
Z-value: 1.60
Transcription factors associated with Smad4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad4
|
ENSMUSG00000024515.7 | SMAD family member 4 |
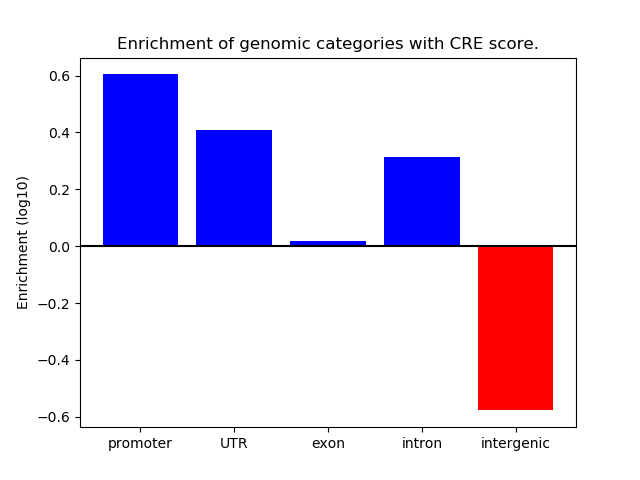
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_73674278_73674444 | Smad4 | 757 | 0.604978 | 0.87 | 2.5e-02 | Click! |
| chr18_73703748_73704065 | Smad4 | 126 | 0.952348 | -0.77 | 7.3e-02 | Click! |
| chr18_73697259_73697432 | Smad4 | 6435 | 0.159697 | 0.77 | 7.5e-02 | Click! |
| chr18_73702216_73702367 | Smad4 | 1489 | 0.339216 | 0.72 | 1.0e-01 | Click! |
| chr18_73697728_73697893 | Smad4 | 5970 | 0.161936 | 0.35 | 5.0e-01 | Click! |
Activity of the Smad4 motif across conditions
Conditions sorted by the z-value of the Smad4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
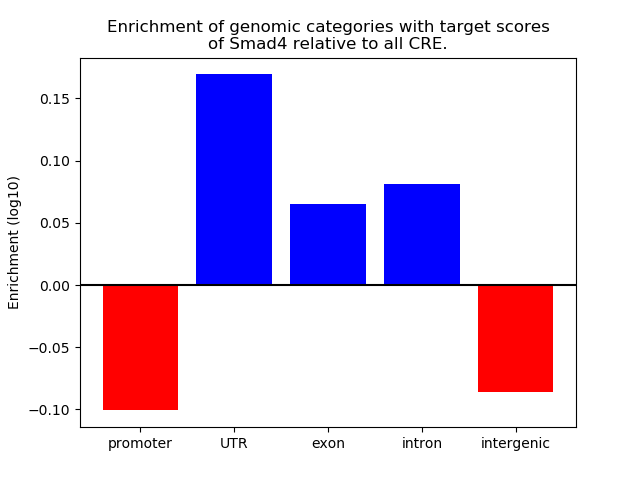
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_35409885_35410137 | 0.97 |
Gm45301 |
predicted gene 45301 |
505 |
0.77 |
| chr15_59020942_59021093 | 0.95 |
Mtss1 |
MTSS I-BAR domain containing 1 |
19579 |
0.18 |
| chr4_120973741_120974003 | 0.93 |
Mir7015 |
microRNA 7015 |
550 |
0.63 |
| chr6_72125643_72125794 | 0.90 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2521 |
0.19 |
| chr19_44397401_44397552 | 0.88 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
9214 |
0.14 |
| chr15_58987050_58987409 | 0.87 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
3093 |
0.22 |
| chr11_72609282_72609433 | 0.85 |
Ube2g1 |
ubiquitin-conjugating enzyme E2G 1 |
2038 |
0.25 |
| chr2_72157539_72157839 | 0.81 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
8893 |
0.2 |
| chr11_60201956_60202762 | 0.80 |
Mir6922 |
microRNA 6922 |
32 |
0.63 |
| chr19_18145861_18146289 | 0.78 |
Gm18610 |
predicted gene, 18610 |
59720 |
0.14 |
| chr12_3647979_3648147 | 0.78 |
Dtnb |
dystrobrevin, beta |
15141 |
0.21 |
| chr13_20660427_20660704 | 0.76 |
Gm47721 |
predicted gene, 47721 |
14808 |
0.21 |
| chr11_16755355_16755534 | 0.75 |
Egfr |
epidermal growth factor receptor |
3214 |
0.26 |
| chr4_131722484_131723038 | 0.74 |
Gm16080 |
predicted gene 16080 |
12253 |
0.19 |
| chr11_63851634_63851785 | 0.74 |
Hmgb1-ps3 |
high mobility group box 1, pseudogene 3 |
4891 |
0.25 |
| chr15_58895415_58895566 | 0.73 |
Rnf139 |
ring finger protein 139 |
4647 |
0.14 |
| chr8_114139321_114139472 | 0.72 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
5754 |
0.3 |
| chr11_75173089_75173918 | 0.71 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr9_122056238_122056617 | 0.71 |
Gm39465 |
predicted gene, 39465 |
4964 |
0.13 |
| chr11_101342858_101343022 | 0.71 |
Gm28156 |
predicted gene 28156 |
4999 |
0.06 |
| chr9_106234042_106234256 | 0.71 |
Alas1 |
aminolevulinic acid synthase 1 |
2935 |
0.14 |
| chrY_90803741_90803892 | 0.70 |
Gm47283 |
predicted gene, 47283 |
13365 |
0.17 |
| chr3_97651056_97651341 | 0.70 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
6995 |
0.14 |
| chr6_23121749_23122234 | 0.70 |
Aass |
aminoadipate-semialdehyde synthase |
5589 |
0.18 |
| chr7_72216332_72216639 | 0.68 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
12875 |
0.25 |
| chr8_93179883_93180034 | 0.68 |
Ces1d |
carboxylesterase 1D |
4669 |
0.15 |
| chr4_117937720_117937882 | 0.66 |
Artn |
artemin |
8038 |
0.12 |
| chr6_143857686_143858000 | 0.66 |
Sox5 |
SRY (sex determining region Y)-box 5 |
89245 |
0.09 |
| chr11_28689224_28689493 | 0.65 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
7794 |
0.19 |
| chr15_3432918_3433085 | 0.63 |
Ghr |
growth hormone receptor |
38643 |
0.2 |
| chr8_42183376_42183557 | 0.62 |
Gm6180 |
predicted pseudogene 6180 |
63445 |
0.14 |
| chr9_106232444_106232608 | 0.61 |
Alas1 |
aminolevulinic acid synthase 1 |
4558 |
0.11 |
| chr12_51274477_51274668 | 0.60 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
22915 |
0.22 |
| chr5_125512652_125512844 | 0.59 |
Aacs |
acetoacetyl-CoA synthetase |
49 |
0.97 |
| chr7_97424078_97424348 | 0.59 |
Thrsp |
thyroid hormone responsive |
6483 |
0.13 |
| chr5_143771106_143771348 | 0.59 |
Gm42503 |
predicted gene 42503 |
1187 |
0.41 |
| chr15_42296635_42296795 | 0.58 |
Gm49452 |
predicted gene, 49452 |
23631 |
0.17 |
| chr3_95881097_95881422 | 0.57 |
Ciart |
circadian associated repressor of transcription |
13 |
0.94 |
| chr5_77314693_77314994 | 0.57 |
Gm42758 |
predicted gene 42758 |
3812 |
0.15 |
| chr14_21195621_21195893 | 0.56 |
Adk |
adenosine kinase |
119605 |
0.05 |
| chr4_118031195_118031493 | 0.56 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
538 |
0.78 |
| chr2_31479250_31479512 | 0.56 |
Ass1 |
argininosuccinate synthetase 1 |
9174 |
0.19 |
| chr19_4594662_4594855 | 0.55 |
Pcx |
pyruvate carboxylase |
350 |
0.8 |
| chr10_18197196_18197617 | 0.53 |
Ect2l |
epithelial cell transforming sequence 2 oncogene-like |
3947 |
0.22 |
| chr9_74993629_74993780 | 0.52 |
Fam214a |
family with sequence similarity 214, member A |
17593 |
0.17 |
| chr4_141958726_141958886 | 0.52 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
4612 |
0.15 |
| chrX_140591571_140591781 | 0.51 |
AL683809.1 |
TSC22 domain family, member 3 (Tsc22d3), pseuodgene |
8077 |
0.17 |
| chr2_73591851_73592008 | 0.51 |
Chn1os3 |
chimerin 1, opposite strand 3 |
4597 |
0.16 |
| chr3_129584585_129584736 | 0.51 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
32280 |
0.14 |
| chr3_28718821_28719132 | 0.51 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
2541 |
0.24 |
| chr4_151043729_151043930 | 0.50 |
Per3 |
period circadian clock 3 |
836 |
0.56 |
| chr11_16818941_16819203 | 0.50 |
Egfros |
epidermal growth factor receptor, opposite strand |
11630 |
0.22 |
| chr7_101344939_101345805 | 0.50 |
Gm45620 |
predicted gene 45620 |
2612 |
0.13 |
| chr19_40180011_40180165 | 0.50 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
7198 |
0.16 |
| chr1_21250942_21251436 | 0.49 |
Gsta3 |
glutathione S-transferase, alpha 3 |
2332 |
0.18 |
| chr11_52350722_52351028 | 0.49 |
Vdac1 |
voltage-dependent anion channel 1 |
9985 |
0.17 |
| chr2_32524318_32524490 | 0.49 |
Gm13412 |
predicted gene 13412 |
627 |
0.55 |
| chr1_21248546_21248733 | 0.49 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4882 |
0.13 |
| chr5_148996162_148996678 | 0.49 |
Gm15406 |
predicted gene 15406 |
89 |
0.9 |
| chr14_30534118_30534505 | 0.48 |
Dcp1a |
decapping mRNA 1A |
10604 |
0.14 |
| chr19_40164076_40164449 | 0.47 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
23024 |
0.14 |
| chrX_75673853_75674412 | 0.46 |
Gm15065 |
predicted gene 15065 |
31277 |
0.13 |
| chr8_121980914_121981086 | 0.46 |
Banp |
BTG3 associated nuclear protein |
2678 |
0.16 |
| chr2_177900583_177900745 | 0.46 |
Gm14327 |
predicted gene 14327 |
3568 |
0.18 |
| chr5_125479676_125479838 | 0.46 |
Gm27551 |
predicted gene, 27551 |
380 |
0.78 |
| chr9_44484103_44484412 | 0.46 |
Gm47205 |
predicted gene, 47205 |
454 |
0.54 |
| chr2_134843502_134843690 | 0.46 |
Gm14036 |
predicted gene 14036 |
39647 |
0.17 |
| chr4_60672835_60672986 | 0.46 |
Mup-ps7 |
major urinary protein, pseudogene 7 |
4773 |
0.19 |
| chr4_99036770_99036925 | 0.45 |
Angptl3 |
angiopoietin-like 3 |
3644 |
0.22 |
| chr5_23562685_23562882 | 0.45 |
Srpk2 |
serine/arginine-rich protein specific kinase 2 |
13413 |
0.19 |
| chr12_104345688_104345839 | 0.45 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
7277 |
0.12 |
| chr12_21302020_21302503 | 0.44 |
Cpsf3 |
cleavage and polyadenylation specificity factor 3 |
990 |
0.4 |
| chr14_58709638_58709806 | 0.44 |
Gm25614 |
predicted gene, 25614 |
59322 |
0.15 |
| chr19_44400900_44401269 | 0.44 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5606 |
0.16 |
| chr8_93176743_93176912 | 0.44 |
Ces1d |
carboxylesterase 1D |
1538 |
0.3 |
| chr1_153741660_153741824 | 0.44 |
Rgs16 |
regulator of G-protein signaling 16 |
1393 |
0.25 |
| chr4_31964283_31964752 | 0.44 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
254 |
0.93 |
| chr6_85811078_85811251 | 0.43 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
1301 |
0.25 |
| chr17_81376267_81376581 | 0.43 |
Gm50044 |
predicted gene, 50044 |
5591 |
0.28 |
| chr11_115905852_115906018 | 0.43 |
Recql5 |
RecQ protein-like 5 |
1318 |
0.24 |
| chr18_77784597_77784781 | 0.43 |
Atp5a1 |
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
4857 |
0.15 |
| chr3_14840901_14841063 | 0.43 |
Car3 |
carbonic anhydrase 3 |
22530 |
0.15 |
| chr16_92610894_92611062 | 0.43 |
Gm26626 |
predicted gene, 26626 |
1836 |
0.39 |
| chr12_104085193_104085357 | 0.43 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
4626 |
0.11 |
| chr17_45999890_46000044 | 0.43 |
Vegfa |
vascular endothelial growth factor A |
21405 |
0.15 |
| chr10_4590008_4590159 | 0.42 |
Gm3266 |
predicted gene 3266 |
19860 |
0.18 |
| chr2_160483998_160484149 | 0.42 |
Gm14222 |
predicted gene 14222 |
99284 |
0.07 |
| chr10_26752927_26753096 | 0.42 |
Arhgap18 |
Rho GTPase activating protein 18 |
410 |
0.83 |
| chrY_90807232_90807393 | 0.42 |
Gm47283 |
predicted gene, 47283 |
16861 |
0.16 |
| chr5_125040097_125040498 | 0.42 |
Ncor2 |
nuclear receptor co-repressor 2 |
2515 |
0.25 |
| chr9_74911852_74912214 | 0.42 |
Fam214a |
family with sequence similarity 214, member A |
40851 |
0.11 |
| chr4_122935638_122935854 | 0.42 |
Gm12891 |
predicted gene 12891 |
9913 |
0.14 |
| chr15_59065322_59065483 | 0.42 |
Mtss1 |
MTSS I-BAR domain containing 1 |
9938 |
0.23 |
| chr13_56440067_56440428 | 0.42 |
Slc25a48 |
solute carrier family 25, member 48 |
1892 |
0.29 |
| chrY_90807930_90808086 | 0.41 |
Gm47283 |
predicted gene, 47283 |
17557 |
0.16 |
| chr1_80244319_80244685 | 0.41 |
Gm37932 |
predicted gene, 37932 |
11695 |
0.13 |
| chr2_31491482_31491666 | 0.41 |
Ass1 |
argininosuccinate synthetase 1 |
6198 |
0.2 |
| chr9_101082169_101082333 | 0.41 |
Msl2 |
MSL complex subunit 2 |
7489 |
0.12 |
| chr11_101291651_101291851 | 0.41 |
Becn1 |
beclin 1, autophagy related |
747 |
0.35 |
| chr3_90081344_90081692 | 0.41 |
Tpm3 |
tropomyosin 3, gamma |
1076 |
0.29 |
| chr9_107328322_107328506 | 0.41 |
Hemk1 |
HemK methyltransferase family member 1 |
1038 |
0.35 |
| chr7_81707314_81707700 | 0.41 |
Homer2 |
homer scaffolding protein 2 |
20 |
0.97 |
| chr1_21264707_21265188 | 0.40 |
Gm28836 |
predicted gene 28836 |
6646 |
0.11 |
| chr3_60525375_60525543 | 0.40 |
Mbnl1 |
muscleblind like splicing factor 1 |
2669 |
0.31 |
| chr16_26669940_26670091 | 0.40 |
Il1rap |
interleukin 1 receptor accessory protein |
45859 |
0.18 |
| chr16_43101819_43101970 | 0.39 |
Gm25873 |
predicted gene, 25873 |
24339 |
0.22 |
| chr13_46052359_46052539 | 0.39 |
Gm45949 |
predicted gene, 45949 |
8148 |
0.26 |
| chr2_147989793_147989944 | 0.39 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
13683 |
0.21 |
| chr5_21737297_21737945 | 0.38 |
Pmpcb |
peptidase (mitochondrial processing) beta |
415 |
0.77 |
| chr2_67898259_67898744 | 0.38 |
Gm37964 |
predicted gene, 37964 |
95 |
0.98 |
| chr14_11803342_11803591 | 0.38 |
Gm48602 |
predicted gene, 48602 |
35523 |
0.19 |
| chr8_120179504_120179692 | 0.38 |
Fam92b |
family with sequence similarity 92, member B |
2132 |
0.25 |
| chr6_85835048_85835221 | 0.38 |
Nat8 |
N-acetyltransferase 8 (GCN5-related) |
3052 |
0.12 |
| chr1_21259850_21260251 | 0.37 |
Gsta3 |
glutathione S-transferase, alpha 3 |
6529 |
0.11 |
| chr1_70918162_70918313 | 0.37 |
Gm16236 |
predicted gene 16236 |
121793 |
0.06 |
| chr11_28698214_28698534 | 0.37 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
16810 |
0.16 |
| chr8_26078485_26078636 | 0.37 |
Hook3 |
hook microtubule tethering protein 3 |
817 |
0.46 |
| chr14_59540860_59541047 | 0.36 |
Cdadc1 |
cytidine and dCMP deaminase domain containing 1 |
56883 |
0.1 |
| chr5_125479434_125479671 | 0.36 |
Gm27551 |
predicted gene, 27551 |
175 |
0.92 |
| chr1_162892140_162892544 | 0.36 |
Fmo2 |
flavin containing monooxygenase 2 |
5827 |
0.19 |
| chr10_123536278_123536442 | 0.36 |
Gm19169 |
predicted gene, 19169 |
47228 |
0.17 |
| chr2_155061918_155062357 | 0.36 |
Gm45609 |
predicted gene 45609 |
12044 |
0.13 |
| chr1_91514960_91515111 | 0.36 |
Traf3ip1 |
TRAF3 interacting protein 1 |
3227 |
0.17 |
| chr12_12793762_12793936 | 0.36 |
Platr19 |
pluripotency associated transcript 19 |
5864 |
0.21 |
| chr7_63916436_63916647 | 0.36 |
E030018B13Rik |
RIKEN cDNA E030018B13 gene |
316 |
0.85 |
| chr6_85763706_85763876 | 0.36 |
Nat8f3 |
N-acetyltransferase 8 (GCN5-related) family member 3 |
1311 |
0.27 |
| chr6_85709780_85709938 | 0.36 |
Nat8f7 |
N-acetyltransferase 8 (GCN5-related) family member 7 |
2001 |
0.2 |
| chr3_138282244_138282429 | 0.36 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
4685 |
0.14 |
| chr7_40677483_40677882 | 0.35 |
4933402C06Rik |
RIKEN cDNA 4933402C06 gene |
2945 |
0.3 |
| chr15_62645080_62645285 | 0.35 |
Gm24810 |
predicted gene, 24810 |
7822 |
0.29 |
| chr2_129865084_129865235 | 0.35 |
Stk35 |
serine/threonine kinase 35 |
64642 |
0.1 |
| chr11_16862611_16863177 | 0.35 |
Egfr |
epidermal growth factor receptor |
15256 |
0.19 |
| chr9_74526287_74526483 | 0.35 |
Gm28622 |
predicted gene 28622 |
34993 |
0.18 |
| chr8_25754579_25754912 | 0.35 |
Ddhd2 |
DDHD domain containing 2 |
149 |
0.63 |
| chr11_105281077_105281256 | 0.35 |
Mrc2 |
mannose receptor, C type 2 |
11477 |
0.19 |
| chr14_29014453_29014621 | 0.35 |
Lrtm1 |
leucine-rich repeats and transmembrane domains 1 |
3671 |
0.23 |
| chr5_102011072_102011265 | 0.35 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
29610 |
0.16 |
| chr7_46727375_46727526 | 0.35 |
Saa4 |
serum amyloid A 4 |
5153 |
0.09 |
| chr10_77441826_77442020 | 0.35 |
Gm35920 |
predicted gene, 35920 |
15994 |
0.15 |
| chr9_44096490_44096809 | 0.35 |
Usp2 |
ubiquitin specific peptidase 2 |
3975 |
0.08 |
| chr14_17823236_17823387 | 0.35 |
Gm48320 |
predicted gene, 48320 |
52189 |
0.17 |
| chr15_59026303_59026454 | 0.35 |
Mtss1 |
MTSS I-BAR domain containing 1 |
14218 |
0.19 |
| chr4_137005965_137006116 | 0.35 |
Zbtb40 |
zinc finger and BTB domain containing 40 |
14378 |
0.16 |
| chr7_46727062_46727213 | 0.34 |
Saa4 |
serum amyloid A 4 |
5466 |
0.09 |
| chr14_19808226_19808383 | 0.34 |
Nid2 |
nidogen 2 |
3068 |
0.21 |
| chr16_93714548_93714944 | 0.34 |
Dop1b |
DOP1 leucine zipper like protein B |
1320 |
0.39 |
| chr5_143526296_143526447 | 0.34 |
Gm42502 |
predicted gene 42502 |
711 |
0.52 |
| chrY_90802780_90802931 | 0.34 |
Gm47283 |
predicted gene, 47283 |
12404 |
0.17 |
| chr15_62329521_62329672 | 0.34 |
Pvt1 |
Pvt1 oncogene |
106993 |
0.07 |
| chr5_102643765_102644161 | 0.34 |
Arhgap24 |
Rho GTPase activating protein 24 |
81010 |
0.11 |
| chr12_69759681_69759832 | 0.34 |
Mir681 |
microRNA 681 |
4188 |
0.15 |
| chr2_58759818_58759990 | 0.34 |
Upp2 |
uridine phosphorylase 2 |
4692 |
0.23 |
| chr9_24502048_24502199 | 0.34 |
Dpy19l1 |
dpy-19-like 1 (C. elegans) |
1017 |
0.45 |
| chr2_178026714_178026865 | 0.34 |
Zfp971 |
zinc finger protein 971 |
3410 |
0.24 |
| chr15_97006005_97006390 | 0.34 |
Slc38a4 |
solute carrier family 38, member 4 |
13754 |
0.26 |
| chr18_12720543_12720734 | 0.34 |
Mir1948 |
microRNA 1948 |
5827 |
0.15 |
| chr7_30475671_30475822 | 0.33 |
Nphs1 |
nephrosis 1, nephrin |
1238 |
0.2 |
| chr13_34076065_34076216 | 0.33 |
Tubb2a |
tubulin, beta 2A class IIA |
1867 |
0.2 |
| chr19_43778712_43778869 | 0.33 |
Abcc2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
3402 |
0.18 |
| chr14_70380431_70380586 | 0.33 |
Gm9174 |
predicted pseudogene 9174 |
20321 |
0.1 |
| chr19_4783214_4783365 | 0.33 |
Rbm4 |
RNA binding motif protein 4 |
10562 |
0.08 |
| chr13_8849368_8849539 | 0.33 |
Wdr37 |
WD repeat domain 37 |
105 |
0.94 |
| chr17_24882806_24883274 | 0.33 |
Nubp2 |
nucleotide binding protein 2 |
2577 |
0.11 |
| chr7_99163511_99163819 | 0.33 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
6441 |
0.13 |
| chr4_122982608_122982816 | 0.33 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
12940 |
0.13 |
| chr9_46099181_46099332 | 0.33 |
Sik3 |
SIK family kinase 3 |
23874 |
0.16 |
| chr1_13587097_13587434 | 0.33 |
Tram1 |
translocating chain-associating membrane protein 1 |
2610 |
0.3 |
| chr1_67178481_67179155 | 0.32 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
55792 |
0.12 |
| chr17_46019375_46019704 | 0.32 |
Vegfa |
vascular endothelial growth factor A |
1833 |
0.31 |
| chr2_71607777_71607946 | 0.32 |
Dlx2 |
distal-less homeobox 2 |
61107 |
0.09 |
| chr9_21248646_21249016 | 0.32 |
S1pr5 |
sphingosine-1-phosphate receptor 5 |
388 |
0.64 |
| chr5_140112771_140112922 | 0.32 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
2620 |
0.23 |
| chr2_31513768_31514291 | 0.32 |
Ass1 |
argininosuccinate synthetase 1 |
4461 |
0.2 |
| chr7_132943127_132943278 | 0.32 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
4966 |
0.16 |
| chr5_125467628_125467779 | 0.32 |
Gm43756 |
predicted gene 43756 |
3794 |
0.14 |
| chr11_5525096_5525390 | 0.32 |
Xbp1 |
X-box binding protein 1 |
3320 |
0.17 |
| chr15_82791292_82791711 | 0.32 |
Cyp2d26 |
cytochrome P450, family 2, subfamily d, polypeptide 26 |
2721 |
0.14 |
| chr3_97649987_97650151 | 0.32 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
8124 |
0.13 |
| chr5_33462368_33462568 | 0.32 |
Gm43851 |
predicted gene 43851 |
25004 |
0.15 |
| chr6_146270994_146271175 | 0.32 |
Gm44086 |
predicted gene, 44086 |
104 |
0.97 |
| chr5_22587487_22587999 | 0.31 |
Gm9057 |
predicted gene 9057 |
23786 |
0.11 |
| chr2_148037591_148038164 | 0.31 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
393 |
0.84 |
| chr9_80672553_80672726 | 0.31 |
Gm39380 |
predicted gene, 39380 |
37356 |
0.19 |
| chr6_53070128_53070316 | 0.31 |
Jazf1 |
JAZF zinc finger 1 |
1591 |
0.41 |
| chr8_61533482_61533653 | 0.31 |
Palld |
palladin, cytoskeletal associated protein |
274 |
0.94 |
| chr1_21252081_21252232 | 0.31 |
Gsta3 |
glutathione S-transferase, alpha 3 |
1365 |
0.29 |
| chr10_96107868_96108070 | 0.31 |
Gm49817 |
predicted gene, 49817 |
11964 |
0.16 |
| chr5_136960851_136961002 | 0.31 |
Fis1 |
fission, mitochondrial 1 |
1151 |
0.29 |
| chr9_35031413_35031583 | 0.31 |
Kirrel3 |
kirre like nephrin family adhesion molecule 3 |
15046 |
0.17 |
| chrY_90799493_90799663 | 0.31 |
Gm47283 |
predicted gene, 47283 |
9127 |
0.18 |
| chr1_41557400_41557596 | 0.30 |
Gm28634 |
predicted gene 28634 |
27955 |
0.26 |
| chr7_70360531_70360745 | 0.30 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
45 |
0.96 |
| chr2_155067847_155068055 | 0.30 |
Gm45609 |
predicted gene 45609 |
6230 |
0.14 |
| chr13_96695184_96695498 | 0.30 |
Gm48575 |
predicted gene, 48575 |
17218 |
0.15 |
| chr3_52011173_52011324 | 0.30 |
Gm37465 |
predicted gene, 37465 |
7223 |
0.13 |
| chr3_94687768_94687919 | 0.30 |
Selenbp2 |
selenium binding protein 2 |
5713 |
0.11 |
| chrY_90800164_90800315 | 0.30 |
Gm47283 |
predicted gene, 47283 |
9788 |
0.18 |
| chr2_130894667_130894834 | 0.30 |
A730017L22Rik |
RIKEN cDNA A730017L22 gene |
11606 |
0.13 |
| chr14_29976665_29976837 | 0.30 |
Actr8 |
ARP8 actin-related protein 8 |
1586 |
0.25 |
| chr18_78777423_78777582 | 0.30 |
Gm23895 |
predicted gene, 23895 |
129374 |
0.05 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 0.6 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.2 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.7 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.4 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.4 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.4 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 0.4 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.1 | 0.2 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.2 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.8 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.3 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.6 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.2 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.2 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.4 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.8 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.2 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.0 | 0.3 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0090181 | regulation of cholesterol metabolic process(GO:0090181) |
| 0.0 | 0.1 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.0 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0043174 | nucleoside salvage(GO:0043174) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.0 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.0 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0051255 | mitotic spindle elongation(GO:0000022) spindle midzone assembly(GO:0051255) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0002141 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.0 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.2 | 0.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0047074 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.3 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.6 | GO:0016749 | N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.5 | GO:0018731 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |