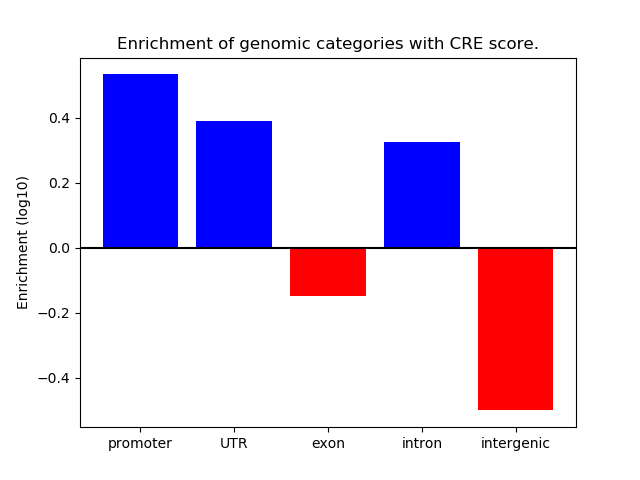
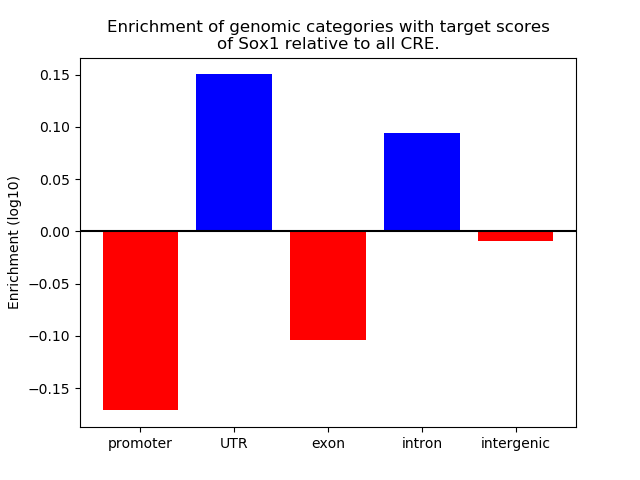
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
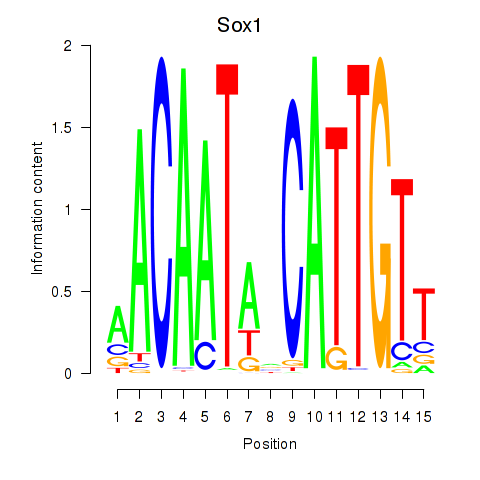
Results for Sox1
Z-value: 0.83
Transcription factors associated with Sox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox1
|
ENSMUSG00000096014.1 | SRY (sex determining region Y)-box 1 |
Activity of the Sox1 motif across conditions
Conditions sorted by the z-value of the Sox1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
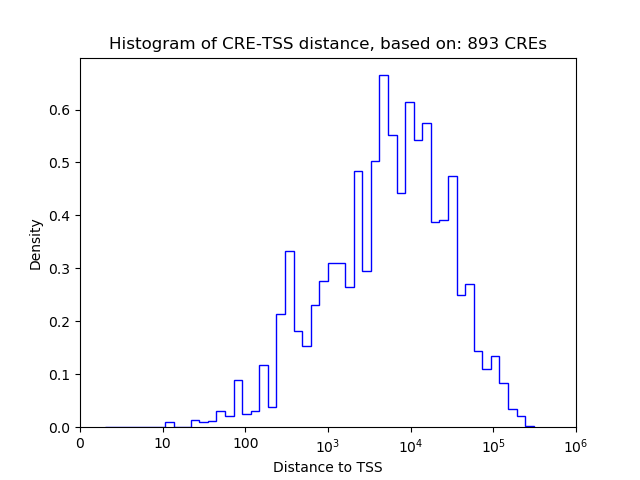
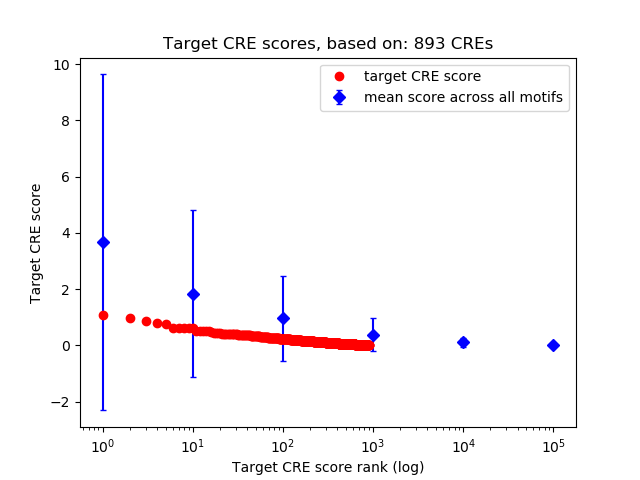
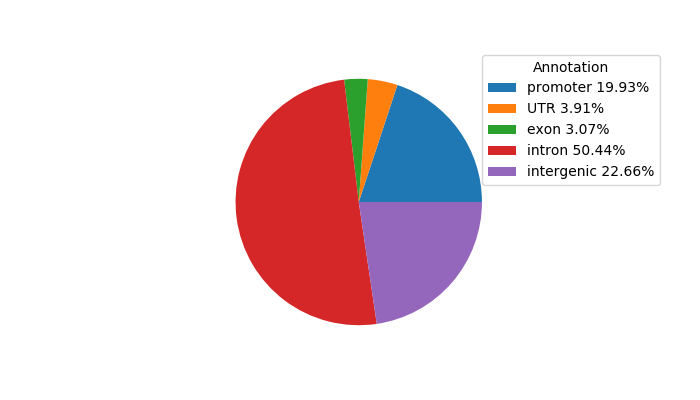
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_16179580_16179731 | 1.09 |
E030024N20Rik |
RIKEN cDNA E030024N20 gene |
14850 |
0.2 |
| chr7_98351056_98351207 | 0.98 |
Tsku |
tsukushi, small leucine rich proteoglycan |
8948 |
0.17 |
| chr19_29520625_29520778 | 0.88 |
A930007I19Rik |
RIKEN cDNA A930007I19 gene |
269 |
0.89 |
| chr10_77011600_77011778 | 0.79 |
Gm10787 |
predicted gene 10787 |
306 |
0.84 |
| chr4_101352284_101352479 | 0.75 |
0610043K17Rik |
RIKEN cDNA 0610043K17 gene |
1402 |
0.26 |
| chr12_80010993_80011167 | 0.62 |
Gm8275 |
predicted gene 8275 |
31229 |
0.14 |
| chr5_86918463_86918614 | 0.61 |
Gm25211 |
predicted gene, 25211 |
2216 |
0.17 |
| chr9_31209643_31210005 | 0.61 |
Aplp2 |
amyloid beta (A4) precursor-like protein 2 |
1963 |
0.34 |
| chr1_85325110_85325261 | 0.60 |
Gm16025 |
predicted gene 16025 |
1544 |
0.27 |
| chr19_44401346_44401497 | 0.60 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5269 |
0.16 |
| chr3_31075182_31075333 | 0.52 |
Skil |
SKI-like |
19801 |
0.16 |
| chr11_90184166_90184514 | 0.51 |
n-R5s72 |
nuclear encoded rRNA 5S 72 |
787 |
0.57 |
| chr14_9492003_9492160 | 0.51 |
Gm48371 |
predicted gene, 48371 |
22526 |
0.26 |
| chr3_112120839_112120990 | 0.51 |
Gm25519 |
predicted gene, 25519 |
78127 |
0.1 |
| chr2_113534474_113534796 | 0.49 |
Gm13964 |
predicted gene 13964 |
30601 |
0.17 |
| chr1_162892544_162892705 | 0.46 |
Fmo2 |
flavin containing monooxygenase 2 |
5860 |
0.19 |
| chr2_71212098_71212533 | 0.45 |
Dync1i2 |
dynein cytoplasmic 1 intermediate chain 2 |
311 |
0.92 |
| chr11_60205641_60205948 | 0.43 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
318 |
0.81 |
| chr19_16029083_16029234 | 0.43 |
Rpl37-ps1 |
ribosomal protein 37, pseudogene 1 |
3707 |
0.24 |
| chr3_88173707_88173903 | 0.43 |
1700113A16Rik |
RIKEN cDNA 1700113A16 gene |
3887 |
0.11 |
| chr4_44976139_44976290 | 0.42 |
Gm12679 |
predicted gene 12679 |
2105 |
0.19 |
| chr5_145454830_145455000 | 0.42 |
Cyp3a16 |
cytochrome P450, family 3, subfamily a, polypeptide 16 |
14808 |
0.16 |
| chr1_74751129_74751319 | 0.41 |
Prkag3 |
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
2003 |
0.21 |
| chr9_103209178_103209657 | 0.41 |
Trf |
transferrin |
2589 |
0.24 |
| chr12_40564154_40564324 | 0.40 |
Dock4 |
dedicator of cytokinesis 4 |
117903 |
0.06 |
| chr1_151339242_151339404 | 0.40 |
Gm10138 |
predicted gene 10138 |
4759 |
0.16 |
| chr7_68140395_68140567 | 0.40 |
Igf1r |
insulin-like growth factor I receptor |
8109 |
0.26 |
| chr12_32764273_32764609 | 0.40 |
Gm18726 |
predicted gene, 18726 |
54280 |
0.11 |
| chr2_31514522_31514710 | 0.39 |
Ass1 |
argininosuccinate synthetase 1 |
3874 |
0.21 |
| chr7_67655501_67655709 | 0.39 |
Ttc23 |
tetratricopeptide repeat domain 23 |
158 |
0.93 |
| chr5_66035556_66035796 | 0.39 |
Rbm47 |
RNA binding motif protein 47 |
18876 |
0.11 |
| chr18_61798684_61798836 | 0.38 |
Afap1l1 |
actin filament associated protein 1-like 1 |
12058 |
0.18 |
| chr3_94699549_94699901 | 0.38 |
Selenbp2 |
selenium binding protein 2 |
6066 |
0.12 |
| chr3_116302483_116302634 | 0.38 |
Gm29151 |
predicted gene 29151 |
47545 |
0.11 |
| chr5_140118402_140118733 | 0.38 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
3101 |
0.21 |
| chr6_149057714_149057872 | 0.37 |
Gm49058 |
predicted gene, 49058 |
3945 |
0.12 |
| chr5_49060180_49060366 | 0.37 |
Gm43047 |
predicted gene 43047 |
9730 |
0.1 |
| chr4_60245923_60246074 | 0.37 |
Mup-ps3 |
major urinary protein, pseudogene 3 |
8653 |
0.19 |
| chr8_45630130_45630742 | 0.37 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
2235 |
0.3 |
| chr17_48484838_48484989 | 0.36 |
Unc5cl |
unc-5 family C-terminal like |
22509 |
0.13 |
| chr4_59006708_59006875 | 0.36 |
Gm20503 |
predicted gene 20503 |
3581 |
0.16 |
| chr9_35209800_35209963 | 0.35 |
Foxred1 |
FAD-dependent oxidoreductase domain containing 1 |
1016 |
0.31 |
| chr4_152264194_152264355 | 0.35 |
Gpr153 |
G protein-coupled receptor 153 |
9958 |
0.12 |
| chr5_117165254_117165405 | 0.35 |
n-R5s174 |
nuclear encoded rRNA 5S 174 |
31226 |
0.11 |
| chr19_10904836_10905155 | 0.35 |
Prpf19 |
pre-mRNA processing factor 19 |
4713 |
0.11 |
| chr4_61167643_61167862 | 0.34 |
Mup-ps9 |
major urinary protein, pseudogene 9 |
4689 |
0.27 |
| chr1_160781397_160781614 | 0.34 |
Rabgap1l |
RAB GTPase activating protein 1-like |
11433 |
0.12 |
| chr9_102354784_102354966 | 0.34 |
Ephb1 |
Eph receptor B1 |
182 |
0.95 |
| chr4_53227508_53227716 | 0.34 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
9755 |
0.17 |
| chr13_3634828_3634979 | 0.34 |
Asb13 |
ankyrin repeat and SOCS box-containing 13 |
804 |
0.58 |
| chr4_61248201_61248394 | 0.33 |
Mup-ps10 |
major urinary protein, pseudogene 10 |
4720 |
0.21 |
| chr13_96648408_96648591 | 0.33 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
4304 |
0.17 |
| chr17_86393949_86394100 | 0.32 |
Prkce |
protein kinase C, epsilon |
96538 |
0.07 |
| chr10_57848460_57848621 | 0.32 |
Smpdl3a |
sphingomyelin phosphodiesterase, acid-like 3A |
53972 |
0.13 |
| chr2_31515291_31515653 | 0.32 |
Ass1 |
argininosuccinate synthetase 1 |
3018 |
0.23 |
| chr15_65337961_65338116 | 0.31 |
Gm49242 |
predicted gene, 49242 |
137476 |
0.05 |
| chr2_26663667_26663830 | 0.31 |
Lcn4 |
lipocalin 4 |
7534 |
0.11 |
| chr3_95885104_95885275 | 0.31 |
Ciart |
circadian associated repressor of transcription |
2938 |
0.1 |
| chr2_163568842_163568993 | 0.31 |
Hnf4a |
hepatic nuclear factor 4, alpha |
18834 |
0.12 |
| chr4_97905915_97906126 | 0.30 |
Nfia |
nuclear factor I/A |
5013 |
0.32 |
| chr7_16598127_16598332 | 0.30 |
Gm29443 |
predicted gene 29443 |
15595 |
0.09 |
| chr10_124590350_124590536 | 0.30 |
4930503E24Rik |
RIKEN cDNA 4930503E24 gene |
67005 |
0.14 |
| chr5_89493650_89493829 | 0.30 |
Gm6366 |
predicted gene 6366 |
6238 |
0.21 |
| chr1_52004207_52004365 | 0.29 |
Stat4 |
signal transducer and activator of transcription 4 |
3957 |
0.2 |
| chr5_87334492_87334755 | 0.29 |
Ugt2a3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
2572 |
0.17 |
| chr2_181319204_181319455 | 0.29 |
Rtel1 |
regulator of telomere elongation helicase 1 |
410 |
0.71 |
| chr14_11774748_11774923 | 0.28 |
Gm48602 |
predicted gene, 48602 |
6892 |
0.28 |
| chr18_31921125_31921293 | 0.28 |
Lims2 |
LIM and senescent cell antigen like domains 2 |
1081 |
0.41 |
| chr17_10417375_10417580 | 0.28 |
A230009B12Rik |
RIKEN cDNA A230009B12 gene |
38739 |
0.18 |
| chr4_86661560_86661711 | 0.28 |
Plin2 |
perilipin 2 |
2487 |
0.26 |
| chr3_133369683_133370155 | 0.27 |
Ppa2 |
pyrophosphatase (inorganic) 2 |
2495 |
0.32 |
| chr7_119940980_119941134 | 0.27 |
Lyrm1 |
LYR motif containing 1 |
4099 |
0.18 |
| chr15_75118840_75119002 | 0.27 |
Ly6c2 |
lymphocyte antigen 6 complex, locus C2 |
6951 |
0.12 |
| chr1_136140658_136140838 | 0.27 |
Kif21b |
kinesin family member 21B |
9294 |
0.11 |
| chr17_34972548_34972711 | 0.27 |
Hspa1l |
heat shock protein 1-like |
74 |
0.8 |
| chr12_70011836_70012241 | 0.27 |
Nin |
ninein |
14789 |
0.11 |
| chr17_80171624_80171842 | 0.27 |
Galm |
galactose mutarotase |
26581 |
0.12 |
| chr7_143757599_143757799 | 0.27 |
Osbpl5 |
oxysterol binding protein-like 5 |
714 |
0.53 |
| chr14_33810284_33810447 | 0.27 |
4930503F20Rik |
RIKEN cDNA 4930503F20 gene |
918 |
0.56 |
| chr10_50881617_50881776 | 0.26 |
Sim1 |
single-minded family bHLH transcription factor 1 |
13058 |
0.25 |
| chr3_98742508_98742984 | 0.26 |
Hsd3b3 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
10719 |
0.13 |
| chr4_107302924_107303085 | 0.26 |
Dio1 |
deiodinase, iodothyronine, type I |
4062 |
0.14 |
| chr10_68134623_68135230 | 0.26 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
1700 |
0.47 |
| chr1_91258114_91258381 | 0.26 |
Ube2f |
ubiquitin-conjugating enzyme E2F (putative) |
6340 |
0.15 |
| chr6_38786978_38787346 | 0.25 |
Hipk2 |
homeodomain interacting protein kinase 2 |
31184 |
0.19 |
| chr2_11568118_11568271 | 0.25 |
Pfkfb3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
14117 |
0.12 |
| chr6_3620394_3620634 | 0.25 |
Vps50 |
VPS50 EARP/GARPII complex subunit |
26496 |
0.17 |
| chr4_92376164_92376339 | 0.25 |
Gm12638 |
predicted gene 12638 |
2903 |
0.4 |
| chr4_137765730_137766012 | 0.25 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
604 |
0.75 |
| chr6_28608968_28609322 | 0.25 |
Gm37978 |
predicted gene, 37978 |
41073 |
0.13 |
| chr14_73776184_73776350 | 0.24 |
Gm24540 |
predicted gene, 24540 |
8299 |
0.18 |
| chr16_26588376_26588753 | 0.24 |
Il1rap |
interleukin 1 receptor accessory protein |
6807 |
0.28 |
| chr2_160665583_160665820 | 0.24 |
Top1 |
topoisomerase (DNA) I |
4432 |
0.21 |
| chr2_104583567_104583832 | 0.24 |
Gm13885 |
predicted gene 13885 |
82 |
0.95 |
| chr11_3136445_3136953 | 0.24 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
692 |
0.58 |
| chr4_102850868_102851049 | 0.24 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
19606 |
0.23 |
| chr3_104424019_104424377 | 0.24 |
Gm9273 |
predicted gene 9273 |
5871 |
0.15 |
| chr1_176797454_176797628 | 0.24 |
Cep170 |
centrosomal protein 170 |
3922 |
0.14 |
| chr5_8164154_8164307 | 0.23 |
Gm21759 |
predicted gene, 21759 |
15406 |
0.17 |
| chr11_110101538_110102129 | 0.23 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
5855 |
0.27 |
| chr8_13158770_13158987 | 0.23 |
Lamp1 |
lysosomal-associated membrane protein 1 |
283 |
0.82 |
| chr9_122865001_122865302 | 0.23 |
Zfp445 |
zinc finger protein 445 |
783 |
0.46 |
| chr4_114987039_114987534 | 0.23 |
Cmpk1 |
cytidine monophosphate (UMP-CMP) kinase 1 |
45 |
0.97 |
| chr7_115795074_115795225 | 0.23 |
Sox6 |
SRY (sex determining region Y)-box 6 |
29561 |
0.24 |
| chr17_65536272_65536447 | 0.23 |
Tmem232 |
transmembrane protein 232 |
4385 |
0.17 |
| chr4_53539908_53540113 | 0.23 |
Slc44a1 |
solute carrier family 44, member 1 |
4398 |
0.24 |
| chr10_3281127_3281278 | 0.22 |
H60c |
histocompatibility 60c |
13431 |
0.18 |
| chr5_21737297_21737945 | 0.22 |
Pmpcb |
peptidase (mitochondrial processing) beta |
415 |
0.77 |
| chr17_86528291_86528457 | 0.22 |
Gm10309 |
predicted gene 10309 |
23142 |
0.2 |
| chr15_103273039_103273393 | 0.22 |
Copz1 |
coatomer protein complex, subunit zeta 1 |
304 |
0.8 |
| chr11_54355641_54355819 | 0.22 |
Gm12224 |
predicted gene 12224 |
2251 |
0.21 |
| chr9_57312626_57312793 | 0.22 |
Gm18996 |
predicted gene, 18996 |
16706 |
0.14 |
| chr15_6209633_6209839 | 0.22 |
Gm23139 |
predicted gene, 23139 |
58388 |
0.13 |
| chr13_41218870_41219122 | 0.21 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
1166 |
0.36 |
| chr15_50267936_50268109 | 0.21 |
Gm49198 |
predicted gene, 49198 |
18876 |
0.3 |
| chr19_24153611_24153973 | 0.21 |
Gm50308 |
predicted gene, 50308 |
278 |
0.9 |
| chr5_87870211_87870412 | 0.21 |
Odam |
odontogenic, ameloblast asssociated |
14718 |
0.1 |
| chr9_54864660_54864839 | 0.21 |
Ireb2 |
iron responsive element binding protein 2 |
960 |
0.53 |
| chr1_36546704_36546886 | 0.21 |
Ankrd39 |
ankyrin repeat domain 39 |
424 |
0.66 |
| chr9_48787148_48787352 | 0.21 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
48695 |
0.15 |
| chr9_69285383_69285592 | 0.20 |
Rora |
RAR-related orphan receptor alpha |
4195 |
0.29 |
| chr10_98666150_98666367 | 0.20 |
Gm5427 |
predicted gene 5427 |
33452 |
0.22 |
| chr9_44391308_44391459 | 0.20 |
Hyou1 |
hypoxia up-regulated 1 |
722 |
0.32 |
| chr1_59176886_59177061 | 0.20 |
Mpp4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
13584 |
0.12 |
| chr12_72490399_72490568 | 0.20 |
Lrrc9 |
leucine rich repeat containing 9 |
5184 |
0.21 |
| chr12_51195911_51196081 | 0.20 |
Gm7172 |
predicted gene 7172 |
61294 |
0.12 |
| chr15_67343180_67343342 | 0.20 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
116492 |
0.06 |
| chr1_130741555_130741763 | 0.20 |
Gm28857 |
predicted gene 28857 |
332 |
0.74 |
| chr18_44525317_44525773 | 0.20 |
Mcc |
mutated in colorectal cancers |
6029 |
0.29 |
| chr5_92725821_92725972 | 0.20 |
Gm20500 |
predicted gene 20500 |
10309 |
0.19 |
| chr13_74019384_74019615 | 0.20 |
Gm6263 |
predicted gene 6263 |
635 |
0.58 |
| chr2_135825531_135825682 | 0.20 |
Gm14210 |
predicted gene 14210 |
12679 |
0.22 |
| chr6_28734300_28734519 | 0.19 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
28689 |
0.19 |
| chr5_8813928_8814079 | 0.19 |
Abcb1b |
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
11287 |
0.15 |
| chr13_80940527_80940956 | 0.19 |
5430425K12Rik |
RIKEN cDNA 5430425K12 gene |
318 |
0.85 |
| chr5_54086048_54086199 | 0.19 |
Gm15820 |
predicted gene 15820 |
8957 |
0.23 |
| chr2_50492868_50493026 | 0.19 |
Gm13488 |
predicted gene 13488 |
28913 |
0.16 |
| chr10_95552026_95552177 | 0.19 |
Mir3058 |
microRNA 3058 |
7220 |
0.13 |
| chr4_138967634_138968170 | 0.19 |
Rnf186 |
ring finger protein 186 |
790 |
0.57 |
| chr3_31045033_31045199 | 0.19 |
Mir7008 |
microRNA 7008 |
6150 |
0.18 |
| chr5_8947059_8947215 | 0.19 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
11415 |
0.13 |
| chr3_118629791_118629942 | 0.19 |
Dpyd |
dihydropyrimidine dehydrogenase |
67680 |
0.11 |
| chr16_43365711_43365882 | 0.19 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1592 |
0.33 |
| chr14_100234504_100234883 | 0.19 |
Gm16260 |
predicted gene 16260 |
15207 |
0.18 |
| chr14_58072759_58072910 | 0.18 |
Fgf9 |
fibroblast growth factor 9 |
148 |
0.96 |
| chr13_18123081_18123244 | 0.18 |
4930448F12Rik |
RIKEN cDNA 4930448F12 gene |
22960 |
0.14 |
| chr11_102852004_102852179 | 0.18 |
Eftud2 |
elongation factor Tu GTP binding domain containing 2 |
4112 |
0.12 |
| chr5_97834854_97835170 | 0.18 |
Antxr2 |
anthrax toxin receptor 2 |
161083 |
0.03 |
| chr9_115342664_115342815 | 0.18 |
Stt3b |
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) |
32318 |
0.11 |
| chr1_164411158_164411348 | 0.18 |
Gm37411 |
predicted gene, 37411 |
13460 |
0.15 |
| chr17_10408684_10408855 | 0.17 |
A230009B12Rik |
RIKEN cDNA A230009B12 gene |
47447 |
0.15 |
| chr9_69317827_69317978 | 0.17 |
Rora |
RAR-related orphan receptor alpha |
28220 |
0.17 |
| chr1_9739980_9740131 | 0.17 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
7593 |
0.15 |
| chr17_51016495_51016646 | 0.17 |
Tbc1d5 |
TBC1 domain family, member 5 |
31944 |
0.23 |
| chr12_55495363_55495542 | 0.17 |
Gm36634 |
predicted gene, 36634 |
2269 |
0.26 |
| chr2_6864030_6864201 | 0.17 |
Celf2 |
CUGBP, Elav-like family member 2 |
7857 |
0.21 |
| chr11_16780614_16780813 | 0.17 |
Egfr |
epidermal growth factor receptor |
28483 |
0.16 |
| chr6_141960851_141961033 | 0.17 |
Slco1a1 |
solute carrier organic anion transporter family, member 1a1 |
13980 |
0.21 |
| chr8_82402195_82403147 | 0.17 |
Il15 |
interleukin 15 |
101 |
0.98 |
| chr6_47533245_47533448 | 0.17 |
Ezh2 |
enhancer of zeste 2 polycomb repressive complex 2 subunit |
1230 |
0.46 |
| chr10_94562316_94562469 | 0.17 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
11520 |
0.17 |
| chr18_68294635_68294920 | 0.17 |
Fam210a |
family with sequence similarity 210, member A |
5425 |
0.17 |
| chr9_65294825_65295003 | 0.16 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
582 |
0.58 |
| chr14_21576956_21577113 | 0.16 |
Kat6b |
K(lysine) acetyltransferase 6B |
45508 |
0.15 |
| chr9_65675680_65676009 | 0.16 |
Oaz2 |
ornithine decarboxylase antizyme 2 |
704 |
0.64 |
| chr7_115674256_115674428 | 0.16 |
Sox6 |
SRY (sex determining region Y)-box 6 |
11977 |
0.31 |
| chr5_109796686_109796837 | 0.16 |
A430073D23Rik |
RIKEN cDNA A430073D23 gene |
463 |
0.79 |
| chr5_89454709_89454919 | 0.16 |
Gc |
vitamin D binding protein |
3084 |
0.3 |
| chr1_182593671_182593859 | 0.16 |
Capn8 |
calpain 8 |
28727 |
0.15 |
| chr7_12949231_12949439 | 0.16 |
1810019N24Rik |
RIKEN cDNA 1810019N24 gene |
731 |
0.41 |
| chr11_60540124_60540296 | 0.16 |
Alkbh5 |
alkB homolog 5, RNA demethylase |
2232 |
0.16 |
| chr1_45924034_45924550 | 0.16 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
970 |
0.47 |
| chr1_161037769_161037920 | 0.16 |
Gas5 |
growth arrest specific 5 |
63 |
0.48 |
| chr5_28056784_28057184 | 0.16 |
Gm26608 |
predicted gene, 26608 |
1525 |
0.35 |
| chr19_47580710_47580962 | 0.16 |
Slk |
STE20-like kinase |
817 |
0.55 |
| chr16_26697784_26698094 | 0.16 |
Il1rap |
interleukin 1 receptor accessory protein |
24495 |
0.23 |
| chr14_25783005_25783218 | 0.16 |
Zcchc24 |
zinc finger, CCHC domain containing 24 |
14072 |
0.14 |
| chr11_55027978_55028300 | 0.16 |
Anxa6 |
annexin A6 |
5276 |
0.18 |
| chr2_75670305_75670456 | 0.15 |
Hnrnpa3 |
heterogeneous nuclear ribonucleoprotein A3 |
7891 |
0.13 |
| chr16_47878967_47879125 | 0.15 |
Gm7275 |
predicted gene 7275 |
193951 |
0.03 |
| chr7_110538178_110538390 | 0.15 |
Gm10087 |
predicted gene 10087 |
11492 |
0.18 |
| chr8_67497857_67498024 | 0.15 |
Nat2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
2992 |
0.21 |
| chr3_102676819_102676970 | 0.15 |
Gm19202 |
predicted gene, 19202 |
9823 |
0.14 |
| chr11_104407376_104407606 | 0.15 |
Gm47315 |
predicted gene, 47315 |
17809 |
0.17 |
| chr3_121439715_121440163 | 0.15 |
Cnn3 |
calponin 3, acidic |
9827 |
0.14 |
| chr10_61606149_61606306 | 0.15 |
Gm28447 |
predicted gene 28447 |
4847 |
0.14 |
| chr15_36421245_36421431 | 0.15 |
Gm49224 |
predicted gene, 49224 |
15353 |
0.14 |
| chr6_114648264_114648681 | 0.15 |
Atg7 |
autophagy related 7 |
374 |
0.9 |
| chr1_86498300_86498465 | 0.15 |
Rpl30-ps6 |
ribosomal protein L30, pseudogene 6 |
24723 |
0.11 |
| chr1_34239606_34239757 | 0.15 |
Dst |
dystonin |
3085 |
0.23 |
| chr3_149224186_149224562 | 0.15 |
Gm10287 |
predicted gene 10287 |
1371 |
0.47 |
| chr1_67235446_67235799 | 0.15 |
Gm15668 |
predicted gene 15668 |
13578 |
0.22 |
| chr2_84649728_84650026 | 0.15 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
397 |
0.71 |
| chr12_84654027_84654189 | 0.15 |
Vrtn |
vertebrae development associated |
11212 |
0.15 |
| chr9_121799539_121799690 | 0.15 |
Hhatl |
hedgehog acyltransferase-like |
7107 |
0.09 |
| chr13_43445030_43445211 | 0.15 |
Ranbp9 |
RAN binding protein 9 |
14324 |
0.13 |
| chr8_81806939_81807093 | 0.14 |
Gm17169 |
predicted gene 17169 |
16108 |
0.2 |
| chr8_46887805_46887956 | 0.14 |
Gm45481 |
predicted gene 45481 |
53968 |
0.1 |
| chr11_16895683_16895855 | 0.14 |
Egfr |
epidermal growth factor receptor |
9416 |
0.2 |
| chr1_60545486_60545637 | 0.14 |
Gm11576 |
predicted gene 11576 |
8541 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.2 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.3 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0097001 | ceramide binding(GO:0097001) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |