Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
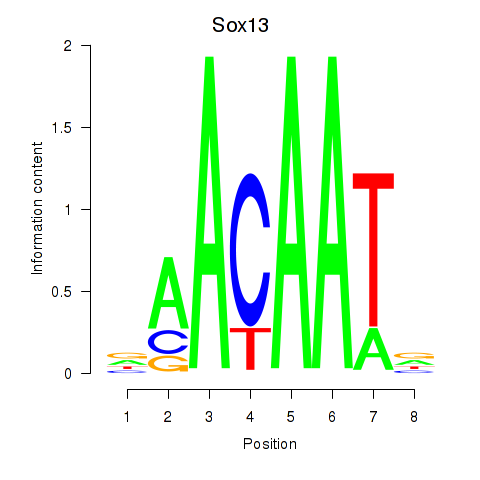
Results for Sox13
Z-value: 1.05
Transcription factors associated with Sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox13
|
ENSMUSG00000070643.5 | SRY (sex determining region Y)-box 13 |
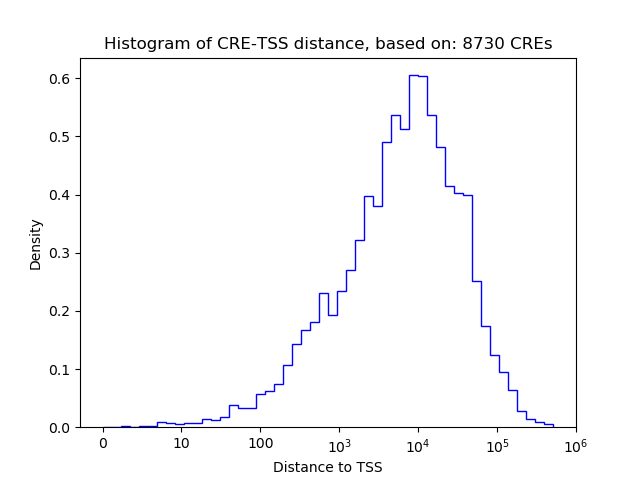
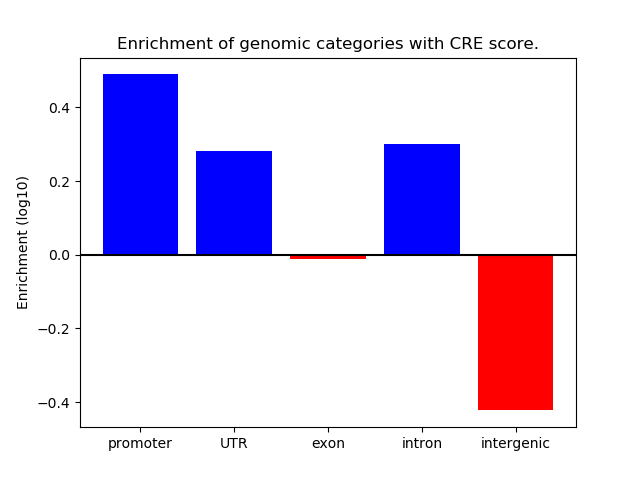
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_133379266_133379589 | Sox13 | 6099 | 0.151951 | 0.83 | 4.0e-02 | Click! |
| chr1_133424091_133424476 | Sox13 | 94 | 0.970159 | -0.79 | 6.0e-02 | Click! |
| chr1_133382772_133383217 | Sox13 | 2532 | 0.220978 | 0.72 | 1.0e-01 | Click! |
| chr1_133452165_133452316 | Sox13 | 27863 | 0.158634 | 0.60 | 2.0e-01 | Click! |
| chr1_133382391_133382543 | Sox13 | 3059 | 0.197220 | 0.60 | 2.0e-01 | Click! |
Activity of the Sox13 motif across conditions
Conditions sorted by the z-value of the Sox13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
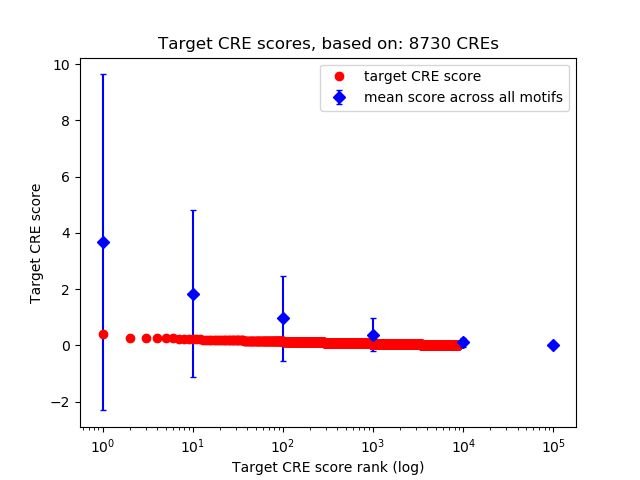
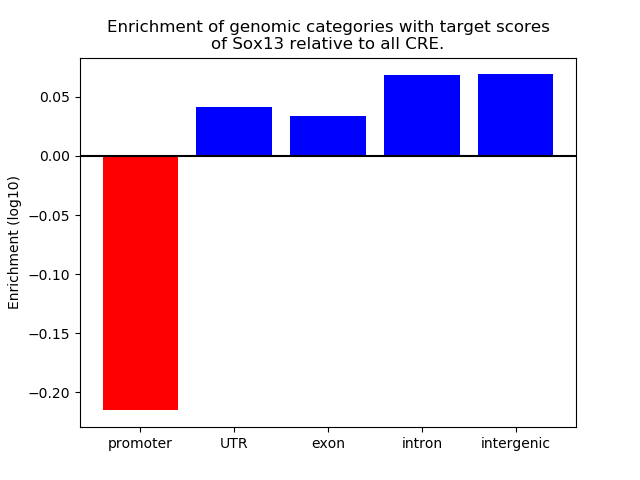
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_25303768_25303919 | 0.40 |
Gm36723 |
predicted gene, 36723 |
97 |
0.92 |
| chr19_17554976_17555127 | 0.26 |
Gm17819 |
predicted gene, 17819 |
137712 |
0.05 |
| chr18_43966502_43966653 | 0.26 |
Spink5 |
serine peptidase inhibitor, Kazal type 5 |
3342 |
0.25 |
| chr19_53781327_53781513 | 0.26 |
Rbm20 |
RNA binding motif protein 20 |
11888 |
0.17 |
| chr2_31519719_31520357 | 0.24 |
Ass1 |
argininosuccinate synthetase 1 |
1548 |
0.36 |
| chr16_24831205_24831356 | 0.24 |
Mir28a |
microRNA 28a |
3425 |
0.28 |
| chr12_16577490_16577750 | 0.22 |
Lpin1 |
lipin 1 |
12100 |
0.23 |
| chr3_116510337_116510525 | 0.22 |
Rtca |
RNA 3'-terminal phosphate cyclase |
2223 |
0.16 |
| chr2_59979696_59979870 | 0.22 |
Gm13574 |
predicted gene 13574 |
12561 |
0.19 |
| chr11_16913067_16913239 | 0.22 |
Egfr |
epidermal growth factor receptor |
7968 |
0.19 |
| chr8_40906070_40906221 | 0.21 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
7707 |
0.17 |
| chr1_184748659_184749028 | 0.21 |
Gm34342 |
predicted gene, 34342 |
11866 |
0.14 |
| chr7_44298836_44299206 | 0.20 |
Gm7238 |
predicted gene 7238 |
1866 |
0.11 |
| chr16_49770118_49770298 | 0.20 |
Gm15518 |
predicted gene 15518 |
28662 |
0.19 |
| chr19_10094218_10094369 | 0.20 |
Fads2 |
fatty acid desaturase 2 |
5577 |
0.16 |
| chr4_95640632_95640805 | 0.20 |
Fggy |
FGGY carbohydrate kinase domain containing |
3831 |
0.31 |
| chr11_99056309_99056460 | 0.20 |
Igfbp4 |
insulin-like growth factor binding protein 4 |
9073 |
0.12 |
| chr10_20504473_20504624 | 0.20 |
Gm17229 |
predicted gene 17229 |
13616 |
0.21 |
| chr11_16845424_16845700 | 0.19 |
Egfros |
epidermal growth factor receptor, opposite strand |
14860 |
0.2 |
| chr12_8498096_8498247 | 0.19 |
Rhob |
ras homolog family member B |
1838 |
0.28 |
| chr4_129063899_129064050 | 0.19 |
Rnf19b |
ring finger protein 19B |
5160 |
0.15 |
| chr2_35174669_35174820 | 0.19 |
Cntrl |
centriolin |
504 |
0.76 |
| chr12_24463865_24464016 | 0.19 |
Gm16372 |
predicted pseudogene 16372 |
29716 |
0.14 |
| chr6_149359539_149359690 | 0.19 |
Gm15784 |
predicted gene 15784 |
2072 |
0.23 |
| chr14_114864274_114864425 | 0.19 |
Gm49010 |
predicted gene, 49010 |
12161 |
0.18 |
| chr2_58737784_58737935 | 0.19 |
Gm13559 |
predicted gene 13559 |
2780 |
0.27 |
| chr7_93588215_93588366 | 0.19 |
Gm45039 |
predicted gene 45039 |
20026 |
0.19 |
| chr13_24359125_24359350 | 0.19 |
Gm11342 |
predicted gene 11342 |
16713 |
0.12 |
| chr19_12460408_12460573 | 0.18 |
Mpeg1 |
macrophage expressed gene 1 |
289 |
0.84 |
| chr3_97620995_97621148 | 0.18 |
Fmo5 |
flavin containing monooxygenase 5 |
7733 |
0.14 |
| chr14_88428338_88428489 | 0.18 |
Gm48930 |
predicted gene, 48930 |
12388 |
0.18 |
| chr19_44399803_44399962 | 0.18 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6808 |
0.15 |
| chrX_108663772_108663923 | 0.18 |
Gm379 |
predicted gene 379 |
608 |
0.84 |
| chr8_109996628_109997116 | 0.17 |
Tat |
tyrosine aminotransferase |
6366 |
0.13 |
| chr2_32415334_32415774 | 0.17 |
Slc25a25 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
1017 |
0.34 |
| chr17_50814204_50814359 | 0.17 |
Gm25177 |
predicted gene, 25177 |
102760 |
0.07 |
| chr4_53222171_53222454 | 0.17 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
4455 |
0.21 |
| chr5_102010547_102010959 | 0.17 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
29195 |
0.16 |
| chr9_100488481_100488638 | 0.17 |
Il20rb |
interleukin 20 receptor beta |
1771 |
0.29 |
| chr3_51238020_51238171 | 0.16 |
Noct |
nocturnin |
2111 |
0.23 |
| chr11_51598115_51598266 | 0.16 |
Phykpl |
5-phosphohydroxy-L-lysine phospholyase |
1077 |
0.39 |
| chr3_122130884_122131070 | 0.16 |
Abca4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
1025 |
0.49 |
| chr7_72295313_72295464 | 0.16 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
11220 |
0.29 |
| chr9_85369091_85369242 | 0.16 |
Gm48832 |
predicted gene, 48832 |
21101 |
0.14 |
| chr11_90269750_90269901 | 0.16 |
Mmd |
monocyte to macrophage differentiation-associated |
20349 |
0.21 |
| chr4_83243559_83243710 | 0.16 |
Ttc39b |
tetratricopeptide repeat domain 39B |
10464 |
0.2 |
| chr2_58784544_58784695 | 0.16 |
Upp2 |
uridine phosphorylase 2 |
19294 |
0.19 |
| chr13_52186671_52186858 | 0.16 |
Gm48199 |
predicted gene, 48199 |
6353 |
0.28 |
| chr4_6270082_6270233 | 0.16 |
Cyp7a1 |
cytochrome P450, family 7, subfamily a, polypeptide 1 |
5474 |
0.21 |
| chr2_43522117_43522300 | 0.16 |
Gm13464 |
predicted gene 13464 |
2810 |
0.36 |
| chr16_85566937_85567088 | 0.16 |
Gm2541 |
predicted pseudogene 2541 |
13069 |
0.22 |
| chr3_158062237_158062399 | 0.16 |
Lrrc40 |
leucine rich repeat containing 40 |
40 |
0.97 |
| chr12_69941545_69941731 | 0.16 |
Atl1 |
atlastin GTPase 1 |
4539 |
0.13 |
| chr14_120368762_120368913 | 0.16 |
Mbnl2 |
muscleblind like splicing factor 2 |
10404 |
0.23 |
| chr18_9782900_9783271 | 0.16 |
Gm23350 |
predicted gene, 23350 |
19823 |
0.12 |
| chr1_67138759_67139000 | 0.16 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
15853 |
0.23 |
| chr18_16665331_16665535 | 0.16 |
Cdh2 |
cadherin 2 |
4636 |
0.31 |
| chr3_81573214_81573380 | 0.16 |
Gm4857 |
predicted gene 4857 |
16472 |
0.28 |
| chr8_36265509_36265688 | 0.15 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
16082 |
0.2 |
| chr18_38888781_38888957 | 0.15 |
Gm5820 |
predicted gene 5820 |
3589 |
0.26 |
| chr19_33489637_33489788 | 0.15 |
Lipo5 |
lipase, member O5 |
16521 |
0.14 |
| chr16_45995065_45995216 | 0.15 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
15078 |
0.15 |
| chr2_58771100_58771251 | 0.15 |
Upp2 |
uridine phosphorylase 2 |
5850 |
0.22 |
| chr3_5396484_5396672 | 0.15 |
4930555M17Rik |
RIKEN cDNA 4930555M17 gene |
118755 |
0.06 |
| chr2_31514522_31514710 | 0.15 |
Ass1 |
argininosuccinate synthetase 1 |
3874 |
0.21 |
| chr11_43688020_43688171 | 0.15 |
Pwwp2a |
PWWP domain containing 2A |
3950 |
0.21 |
| chr14_116316024_116316184 | 0.15 |
Gm38045 |
predicted gene, 38045 |
434111 |
0.01 |
| chr10_21665548_21665708 | 0.15 |
Gm5420 |
predicted gene 5420 |
20783 |
0.2 |
| chrY_90734416_90734573 | 0.15 |
Mid1-ps1 |
midline 1, pseudogene 1 |
18563 |
0.17 |
| chr3_18152374_18152525 | 0.15 |
Gm23686 |
predicted gene, 23686 |
25176 |
0.2 |
| chr16_87670516_87670809 | 0.14 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
28283 |
0.17 |
| chr4_150868400_150868675 | 0.14 |
Errfi1 |
ERBB receptor feedback inhibitor 1 |
13464 |
0.13 |
| chr1_67210897_67211317 | 0.14 |
Gm15668 |
predicted gene 15668 |
38093 |
0.16 |
| chr2_58779426_58779577 | 0.14 |
Upp2 |
uridine phosphorylase 2 |
14176 |
0.2 |
| chr5_33024447_33024632 | 0.14 |
Ywhah |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
2748 |
0.21 |
| chr3_104218648_104218830 | 0.14 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
1364 |
0.29 |
| chr2_134853818_134853969 | 0.14 |
Gm14036 |
predicted gene 14036 |
49944 |
0.15 |
| chr4_62100969_62101120 | 0.14 |
Gm12910 |
predicted gene 12910 |
11887 |
0.13 |
| chr8_93186224_93186375 | 0.14 |
Gm45909 |
predicted gene 45909 |
5059 |
0.15 |
| chr2_147998244_147998434 | 0.14 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
22154 |
0.18 |
| chrX_169972936_169973308 | 0.14 |
Mid1 |
midline 1 |
237 |
0.91 |
| chr17_28420256_28420436 | 0.14 |
Fkbp5 |
FK506 binding protein 5 |
1819 |
0.21 |
| chr8_67497857_67498024 | 0.14 |
Nat2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
2992 |
0.21 |
| chr7_119850861_119851025 | 0.14 |
Rexo5 |
RNA exonuclease 5 |
6737 |
0.15 |
| chr5_130017444_130017684 | 0.14 |
Asl |
argininosuccinate lyase |
2842 |
0.16 |
| chr11_61612422_61612587 | 0.14 |
Epn2 |
epsin 2 |
32817 |
0.12 |
| chr6_23115074_23115260 | 0.14 |
Aass |
aminoadipate-semialdehyde synthase |
12413 |
0.16 |
| chr7_136493976_136494136 | 0.14 |
Gm36849 |
predicted gene, 36849 |
140692 |
0.04 |
| chr1_67156793_67157207 | 0.14 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
33974 |
0.18 |
| chr7_125056317_125056468 | 0.14 |
Gm45093 |
predicted gene 45093 |
13099 |
0.23 |
| chr12_20815638_20815951 | 0.14 |
1700030C10Rik |
RIKEN cDNA 1700030C10 gene |
15 |
0.98 |
| chr1_52465382_52465533 | 0.14 |
Nab1 |
Ngfi-A binding protein 1 |
1600 |
0.34 |
| chr1_186581211_186581378 | 0.14 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
22613 |
0.2 |
| chr10_87565564_87566020 | 0.14 |
Pah |
phenylalanine hydroxylase |
19119 |
0.21 |
| chr1_24116155_24116306 | 0.14 |
Gm26607 |
predicted gene, 26607 |
9045 |
0.17 |
| chr11_16869435_16869808 | 0.14 |
Egfr |
epidermal growth factor receptor |
8529 |
0.21 |
| chr19_40141969_40142120 | 0.14 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
45242 |
0.11 |
| chr19_47578801_47579105 | 0.14 |
Slk |
STE20-like kinase |
725 |
0.6 |
| chr7_101084310_101084461 | 0.14 |
Fchsd2 |
FCH and double SH3 domains 2 |
8478 |
0.17 |
| chr13_4234942_4235096 | 0.14 |
Akr1c19 |
aldo-keto reductase family 1, member C19 |
1279 |
0.37 |
| chr17_55903580_55903737 | 0.14 |
Zfp959 |
zinc finger protein 959 |
11190 |
0.1 |
| chr11_98192679_98192909 | 0.13 |
Med1 |
mediator complex subunit 1 |
161 |
0.92 |
| chr2_31499430_31499808 | 0.13 |
Ass1 |
argininosuccinate synthetase 1 |
1107 |
0.5 |
| chr6_95118485_95118647 | 0.13 |
Kbtbd8 |
kelch repeat and BTB (POZ) domain containing 8 |
582 |
0.58 |
| chr6_66996154_66996403 | 0.13 |
Gm36816 |
predicted gene, 36816 |
12135 |
0.12 |
| chr5_38117047_38117323 | 0.13 |
Stx18 |
syntaxin 18 |
3724 |
0.2 |
| chr7_118572136_118572408 | 0.13 |
Tmc7 |
transmembrane channel-like gene family 7 |
5645 |
0.16 |
| chr10_24258031_24258221 | 0.13 |
Moxd1 |
monooxygenase, DBH-like 1 |
34581 |
0.16 |
| chr7_49734781_49734959 | 0.13 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
24245 |
0.19 |
| chr19_33480626_33480777 | 0.13 |
Lipo5 |
lipase, member O5 |
7510 |
0.16 |
| chr18_38856050_38856246 | 0.13 |
Fgf1 |
fibroblast growth factor 1 |
2600 |
0.31 |
| chr5_148633016_148633167 | 0.13 |
Gm29815 |
predicted gene, 29815 |
19447 |
0.18 |
| chr10_55194027_55194219 | 0.13 |
Gm46190 |
predicted gene, 46190 |
69761 |
0.14 |
| chr5_137975596_137975772 | 0.13 |
Azgp1 |
alpha-2-glycoprotein 1, zinc |
5836 |
0.09 |
| chr8_56345237_56345388 | 0.13 |
Gm45540 |
predicted gene 45540 |
34008 |
0.18 |
| chr13_114485155_114485339 | 0.13 |
Fst |
follistatin |
26296 |
0.14 |
| chr6_42251414_42251583 | 0.13 |
Gstk1 |
glutathione S-transferase kappa 1 |
5475 |
0.12 |
| chr3_51233010_51233232 | 0.13 |
Gm38357 |
predicted gene, 38357 |
1204 |
0.38 |
| chr3_109307549_109307748 | 0.13 |
Vav3 |
vav 3 oncogene |
33005 |
0.21 |
| chr1_121297700_121297851 | 0.13 |
Gm38283 |
predicted gene, 38283 |
3056 |
0.23 |
| chr10_69208679_69208830 | 0.13 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
202 |
0.94 |
| chr11_16855071_16855379 | 0.13 |
Egfr |
epidermal growth factor receptor |
22925 |
0.17 |
| chr1_73966648_73966904 | 0.13 |
Tns1 |
tensin 1 |
3733 |
0.28 |
| chr9_53288790_53288968 | 0.13 |
Exph5 |
exophilin 5 |
12791 |
0.18 |
| chr7_116331137_116331288 | 0.13 |
Rps13 |
ribosomal protein S13 |
1970 |
0.19 |
| chr19_6053827_6053978 | 0.13 |
AC131692.1 |
mitochondrial ribosomal protein L49 (Mrpl49) pseudogene |
1764 |
0.1 |
| chr16_46841628_46841787 | 0.13 |
Gm6912 |
predicted gene 6912 |
232491 |
0.02 |
| chr16_24201028_24201179 | 0.13 |
Gm31814 |
predicted gene, 31814 |
15143 |
0.2 |
| chr3_9751433_9751584 | 0.13 |
Gm16337 |
predicted gene 16337 |
6099 |
0.23 |
| chr9_14045065_14045224 | 0.13 |
1700019J19Rik |
RIKEN cDNA 1700019J19 gene |
44476 |
0.14 |
| chr14_62001768_62001919 | 0.13 |
Gm47953 |
predicted gene, 47953 |
10476 |
0.25 |
| chr8_10899350_10899661 | 0.13 |
4833411C07Rik |
RIKEN cDNA 4833411C07 gene |
417 |
0.64 |
| chr19_10062797_10062948 | 0.12 |
Fads3 |
fatty acid desaturase 3 |
6620 |
0.14 |
| chr10_40511211_40511581 | 0.12 |
Gm18671 |
predicted gene, 18671 |
37496 |
0.14 |
| chr11_63884359_63884556 | 0.12 |
Hmgb1-ps3 |
high mobility group box 1, pseudogene 3 |
37639 |
0.15 |
| chr2_45463913_45464105 | 0.12 |
Gm13479 |
predicted gene 13479 |
106744 |
0.07 |
| chr2_58783629_58783834 | 0.12 |
Upp2 |
uridine phosphorylase 2 |
18406 |
0.19 |
| chrY_90810953_90811113 | 0.12 |
Gm47283 |
predicted gene, 47283 |
20582 |
0.16 |
| chr11_7815566_7815774 | 0.12 |
Gm27393 |
predicted gene, 27393 |
70835 |
0.13 |
| chr14_86309035_86309286 | 0.12 |
Gm23926 |
predicted gene, 23926 |
2218 |
0.22 |
| chr10_10818225_10818395 | 0.12 |
4930567K20Rik |
RIKEN cDNA 4930567K20 gene |
67230 |
0.1 |
| chr11_16889902_16890060 | 0.12 |
Egfr |
epidermal growth factor receptor |
11831 |
0.19 |
| chr17_64609780_64609995 | 0.12 |
Man2a1 |
mannosidase 2, alpha 1 |
9151 |
0.26 |
| chr16_25011043_25011318 | 0.12 |
A230028O05Rik |
RIKEN cDNA A230028O05 gene |
48459 |
0.18 |
| chr3_141502934_141503090 | 0.12 |
Unc5c |
unc-5 netrin receptor C |
37341 |
0.18 |
| chr14_11801019_11801346 | 0.12 |
Gm48602 |
predicted gene, 48602 |
33239 |
0.19 |
| chr5_125325436_125325605 | 0.12 |
Scarb1 |
scavenger receptor class B, member 1 |
747 |
0.57 |
| chrY_90794490_90794856 | 0.12 |
Gm47283 |
predicted gene, 47283 |
4222 |
0.21 |
| chr13_101895557_101895722 | 0.12 |
Gm17832 |
predicted gene, 17832 |
24781 |
0.19 |
| chr1_193926856_193927030 | 0.12 |
Gm21362 |
predicted gene, 21362 |
59934 |
0.16 |
| chr10_4599676_4599928 | 0.12 |
Esr1 |
estrogen receptor 1 (alpha) |
11791 |
0.21 |
| chr3_52618299_52618553 | 0.12 |
Gm10293 |
predicted pseudogene 10293 |
5591 |
0.27 |
| chr6_43851681_43851832 | 0.12 |
Gm7783 |
predicted gene 7783 |
5148 |
0.27 |
| chr19_44398583_44398737 | 0.12 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
8030 |
0.15 |
| chr5_100371529_100371680 | 0.12 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
1880 |
0.3 |
| chr5_135713964_135714115 | 0.12 |
Por |
P450 (cytochrome) oxidoreductase |
1966 |
0.2 |
| chr18_43980831_43980982 | 0.12 |
Spink5 |
serine peptidase inhibitor, Kazal type 5 |
17671 |
0.17 |
| chr13_46023839_46023999 | 0.12 |
Gm45949 |
predicted gene, 45949 |
36678 |
0.16 |
| chr8_3493535_3493936 | 0.12 |
Zfp358 |
zinc finger protein 358 |
565 |
0.6 |
| chr8_23189899_23190050 | 0.12 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
964 |
0.41 |
| chr11_29526849_29527005 | 0.12 |
Mtif2 |
mitochondrial translational initiation factor 2 |
470 |
0.69 |
| chr10_68093593_68093917 | 0.12 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
42871 |
0.14 |
| chr4_99028977_99029128 | 0.12 |
Angptl3 |
angiopoietin-like 3 |
1902 |
0.32 |
| chr16_22511358_22511522 | 0.12 |
9230117E06Rik |
RIKEN cDNA 9230117E06 gene |
40 |
0.96 |
| chr2_38585174_38585360 | 0.12 |
Gm44183 |
predicted gene, 44183 |
18913 |
0.12 |
| chr19_46603380_46603531 | 0.12 |
Wbp1l |
WW domain binding protein 1 like |
4331 |
0.16 |
| chr5_87499011_87499178 | 0.12 |
Ugt2a1 |
UDP glucuronosyltransferase 2 family, polypeptide A1 |
8223 |
0.12 |
| chr6_31110022_31110173 | 0.12 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
823 |
0.49 |
| chr11_16883025_16883203 | 0.12 |
Egfr |
epidermal growth factor receptor |
4964 |
0.23 |
| chr3_107315148_107315299 | 0.12 |
Rbm15 |
RNA binding motif protein 15 |
17878 |
0.13 |
| chr13_63231532_63231825 | 0.12 |
Aopep |
aminopeptidase O |
8351 |
0.11 |
| chr4_45514779_45514987 | 0.12 |
Gm22518 |
predicted gene, 22518 |
14331 |
0.14 |
| chr15_97047820_97047997 | 0.12 |
Slc38a4 |
solute carrier family 38, member 4 |
2179 |
0.42 |
| chr10_10925217_10925440 | 0.12 |
Gm48406 |
predicted gene, 48406 |
110588 |
0.06 |
| chr2_59979903_59980091 | 0.12 |
Gm13574 |
predicted gene 13574 |
12347 |
0.19 |
| chr8_40875940_40876218 | 0.12 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
1131 |
0.44 |
| chr17_27621069_27621255 | 0.12 |
Nudt3 |
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
1231 |
0.25 |
| chr2_32680438_32680786 | 0.11 |
Fpgs |
folylpolyglutamyl synthetase |
4392 |
0.08 |
| chr5_20881136_20881540 | 0.11 |
Phtf2 |
putative homeodomain transcription factor 2 |
707 |
0.46 |
| chr8_40882534_40882732 | 0.11 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
7685 |
0.16 |
| chr19_10052563_10053085 | 0.11 |
Fads3 |
fatty acid desaturase 3 |
127 |
0.94 |
| chr10_52234541_52234692 | 0.11 |
Dcbld1 |
discoidin, CUB and LCCL domain containing 1 |
979 |
0.54 |
| chr2_115537784_115537935 | 0.11 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
10115 |
0.22 |
| chr19_58370035_58370186 | 0.11 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
84356 |
0.09 |
| chr1_184276149_184276321 | 0.11 |
Gm37223 |
predicted gene, 37223 |
82094 |
0.09 |
| chr1_89710237_89710633 | 0.11 |
4933400F21Rik |
RIKEN cDNA 4933400F21 gene |
26527 |
0.21 |
| chr3_116302483_116302634 | 0.11 |
Gm29151 |
predicted gene 29151 |
47545 |
0.11 |
| chr13_114469994_114470145 | 0.11 |
Fst |
follistatin |
11118 |
0.15 |
| chr12_103768009_103768212 | 0.11 |
Serpina1d |
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
5482 |
0.12 |
| chr10_19457260_19457442 | 0.11 |
Gm33104 |
predicted gene, 33104 |
29364 |
0.18 |
| chr10_4531024_4531190 | 0.11 |
Ccdc170 |
coiled-coil domain containing 170 |
1888 |
0.34 |
| chr11_99081665_99081816 | 0.11 |
Tns4 |
tensin 4 |
6399 |
0.15 |
| chr1_58190709_58190860 | 0.11 |
Aox4 |
aldehyde oxidase 4 |
19613 |
0.16 |
| chr8_54468376_54468527 | 0.11 |
Gm45553 |
predicted gene 45553 |
45699 |
0.14 |
| chr18_75499322_75499625 | 0.11 |
Gm10532 |
predicted gene 10532 |
15172 |
0.25 |
| chr13_51089168_51089510 | 0.11 |
Spin1 |
spindlin 1 |
11541 |
0.24 |
| chr1_136893539_136893740 | 0.11 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
46944 |
0.13 |
| chr17_80090476_80090673 | 0.11 |
Gm23125 |
predicted gene, 23125 |
14363 |
0.14 |
| chr16_36770901_36771058 | 0.11 |
Slc15a2 |
solute carrier family 15 (H+/peptide transporter), member 2 |
2299 |
0.18 |
| chr5_20858729_20858886 | 0.11 |
Phtf2 |
putative homeodomain transcription factor 2 |
3616 |
0.22 |
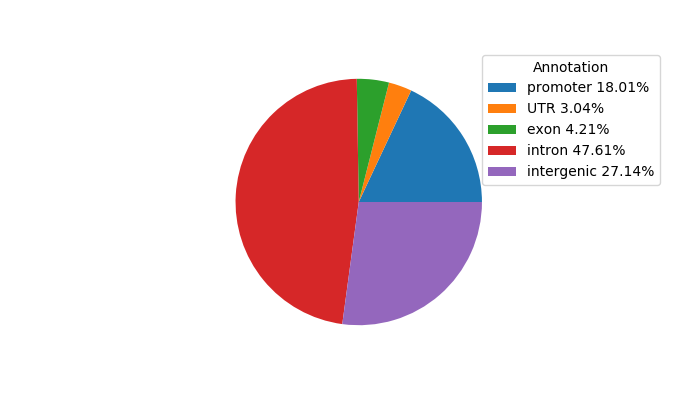
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.2 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |