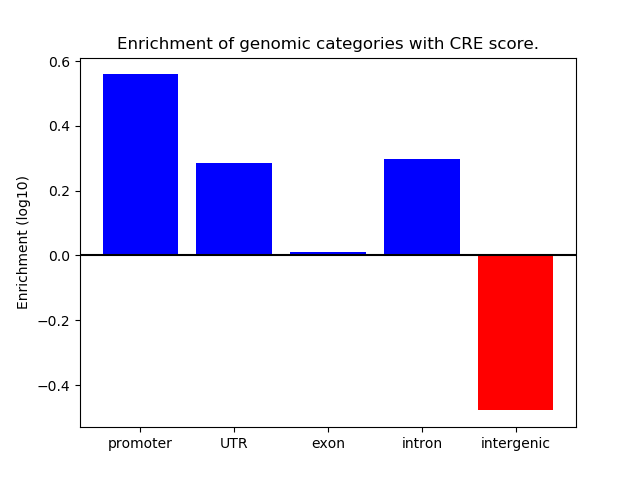
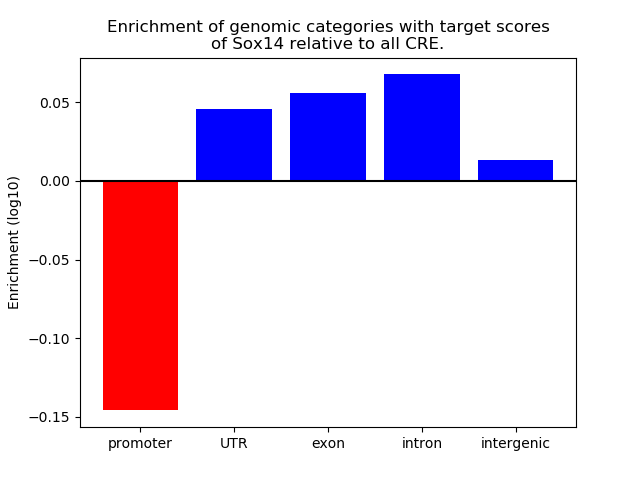
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
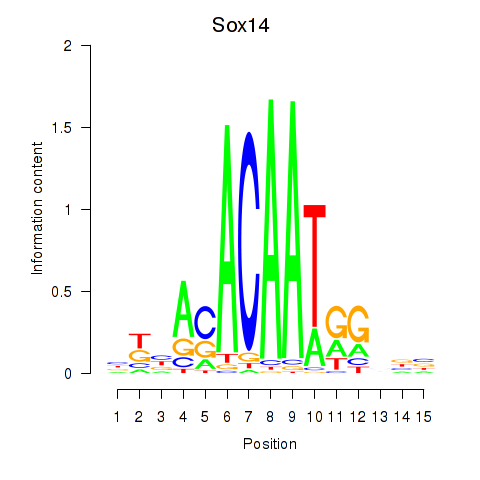
Results for Sox14
Z-value: 1.45
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSMUSG00000053747.8 | SRY (sex determining region Y)-box 14 |
Activity of the Sox14 motif across conditions
Conditions sorted by the z-value of the Sox14 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
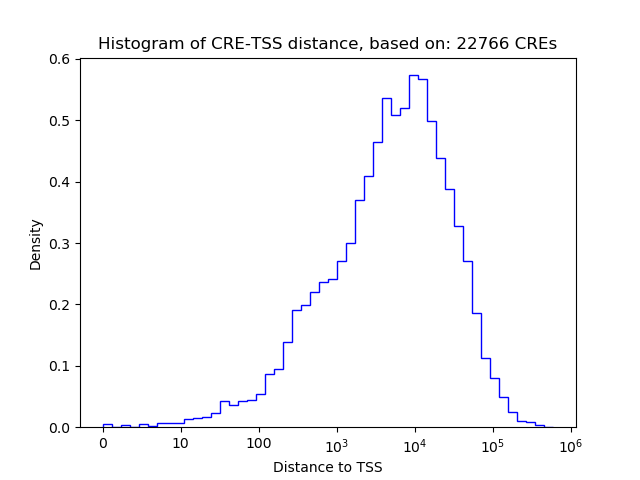
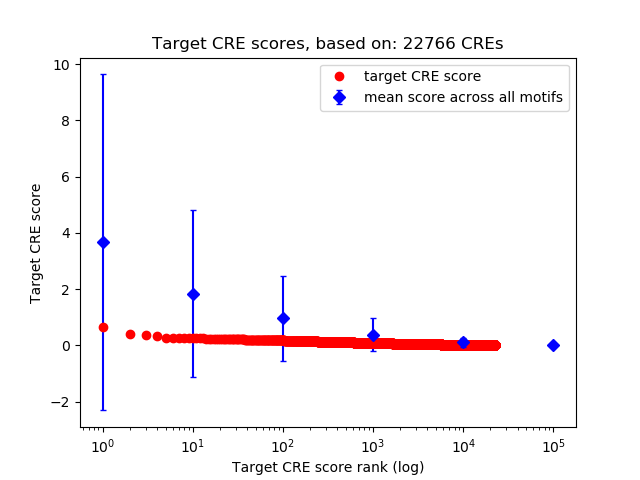
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_158062237_158062399 | 0.67 |
Lrrc40 |
leucine rich repeat containing 40 |
40 |
0.97 |
| chr3_138289329_138289480 | 0.41 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
11753 |
0.11 |
| chr9_9565147_9565311 | 0.35 |
Gm47334 |
predicted gene, 47334 |
1142 |
0.57 |
| chr17_57213268_57213419 | 0.31 |
Mir6978 |
microRNA 6978 |
3885 |
0.13 |
| chr6_108811508_108811751 | 0.27 |
Arl8b |
ADP-ribosylation factor-like 8B |
16732 |
0.19 |
| chr2_121415270_121415468 | 0.26 |
Pdia3 |
protein disulfide isomerase associated 3 |
1261 |
0.21 |
| chr14_65783730_65783949 | 0.25 |
Pbk |
PDZ binding kinase |
21998 |
0.18 |
| chr15_66609811_66609974 | 0.25 |
Phf20l1 |
PHD finger protein 20-like 1 |
4202 |
0.26 |
| chr4_82470714_82470939 | 0.25 |
Nfib |
nuclear factor I/B |
28490 |
0.17 |
| chr17_31904457_31904633 | 0.25 |
2310015A16Rik |
RIKEN cDNA 2310015A16 gene |
4484 |
0.15 |
| chr2_71607065_71607232 | 0.25 |
Dlx2 |
distal-less homeobox 2 |
60394 |
0.09 |
| chr16_36075880_36076031 | 0.25 |
Ccdc58 |
coiled-coil domain containing 58 |
3871 |
0.13 |
| chr2_84757629_84757780 | 0.24 |
Gm19426 |
predicted gene, 19426 |
14049 |
0.08 |
| chr10_84418574_84418728 | 0.24 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
21946 |
0.15 |
| chr8_93195822_93196100 | 0.23 |
Ces1d |
carboxylesterase 1D |
1816 |
0.25 |
| chr6_149359539_149359690 | 0.23 |
Gm15784 |
predicted gene 15784 |
2072 |
0.23 |
| chr11_16779369_16779559 | 0.23 |
Egfr |
epidermal growth factor receptor |
27234 |
0.17 |
| chr10_69205623_69205774 | 0.23 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2854 |
0.25 |
| chr13_73768583_73768734 | 0.23 |
Slc12a7 |
solute carrier family 12, member 7 |
4919 |
0.19 |
| chr1_134562594_134562745 | 0.23 |
Kdm5b |
lysine (K)-specific demethylase 5B |
2462 |
0.2 |
| chr7_119671311_119671666 | 0.23 |
Acsm4 |
acyl-CoA synthetase medium-chain family member 4 |
18538 |
0.1 |
| chr1_5019068_5019219 | 0.23 |
Rgs20 |
regulator of G-protein signaling 20 |
236 |
0.92 |
| chr19_17761902_17762135 | 0.22 |
Gm17819 |
predicted gene, 17819 |
69255 |
0.1 |
| chr3_129451172_129451350 | 0.22 |
Rpl7a-ps7 |
ribosomal protein L7A, pseudogene 7 |
10711 |
0.17 |
| chr5_120481198_120481714 | 0.22 |
Gm15690 |
predicted gene 15690 |
553 |
0.54 |
| chr7_98352590_98353259 | 0.22 |
Tsku |
tsukushi, small leucine rich proteoglycan |
7155 |
0.18 |
| chr2_86337007_86337158 | 0.22 |
Olfr1054 |
olfactory receptor 1054 |
1649 |
0.14 |
| chr2_51145289_51145440 | 0.22 |
Rnd3 |
Rho family GTPase 3 |
3730 |
0.3 |
| chr2_73940118_73940281 | 0.22 |
Gm13668 |
predicted gene 13668 |
20470 |
0.18 |
| chr2_60940502_60940685 | 0.21 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
22599 |
0.21 |
| chr2_38823684_38823849 | 0.21 |
Nr6a1os |
nuclear receptor subfamily 6, group A, member 1, opposite strand |
425 |
0.74 |
| chr8_93188530_93188681 | 0.21 |
Gm45909 |
predicted gene 45909 |
2753 |
0.19 |
| chr14_30894510_30894661 | 0.21 |
Itih4 |
inter alpha-trypsin inhibitor, heavy chain 4 |
4576 |
0.13 |
| chr5_29584226_29584582 | 0.21 |
Ube3c |
ubiquitin protein ligase E3C |
6425 |
0.21 |
| chr1_3424638_3424789 | 0.21 |
Gm1992 |
predicted gene 1992 |
41874 |
0.12 |
| chr6_111333846_111334035 | 0.21 |
Grm7 |
glutamate receptor, metabotropic 7 |
161775 |
0.04 |
| chr11_16798866_16799017 | 0.21 |
Egfros |
epidermal growth factor receptor, opposite strand |
31761 |
0.16 |
| chr16_26619267_26619418 | 0.21 |
Il1rap |
interleukin 1 receptor accessory protein |
4814 |
0.32 |
| chr4_24574389_24574545 | 0.21 |
Mms22l |
MMS22-like, DNA repair protein |
4381 |
0.32 |
| chr8_46497442_46498284 | 0.21 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
5031 |
0.18 |
| chr10_68266351_68266502 | 0.20 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
12295 |
0.22 |
| chr4_101352284_101352479 | 0.20 |
0610043K17Rik |
RIKEN cDNA 0610043K17 gene |
1402 |
0.26 |
| chr6_95710009_95710160 | 0.20 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
8716 |
0.32 |
| chr2_164008768_164008921 | 0.20 |
Ywhab |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
13341 |
0.15 |
| chr9_77743734_77743885 | 0.20 |
Gm47795 |
predicted gene, 47795 |
10541 |
0.13 |
| chr4_144974012_144974163 | 0.20 |
Vps13d |
vacuolar protein sorting 13D |
9874 |
0.2 |
| chr2_166989507_166989658 | 0.20 |
Stau1 |
staufen double-stranded RNA binding protein 1 |
6521 |
0.12 |
| chr14_30573310_30573517 | 0.20 |
Mir3076 |
microRNA 3076 |
1264 |
0.37 |
| chr9_122125893_122126094 | 0.19 |
4632418H02Rik |
RIKEN cDNA 4632418H02 gene |
1301 |
0.32 |
| chr4_63057046_63057197 | 0.19 |
Zfp618 |
zinc finger protein 618 |
63279 |
0.1 |
| chr14_17725181_17725373 | 0.19 |
Gm48320 |
predicted gene, 48320 |
45845 |
0.18 |
| chr18_43132120_43132271 | 0.19 |
Gm8181 |
predicted gene 8181 |
38964 |
0.13 |
| chr9_119148992_119149143 | 0.19 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
996 |
0.4 |
| chr15_88858344_88858998 | 0.19 |
Pim3 |
proviral integration site 3 |
3515 |
0.17 |
| chrX_140506043_140506236 | 0.19 |
Tsc22d3 |
TSC22 domain family, member 3 |
36529 |
0.14 |
| chr13_42723484_42723635 | 0.19 |
Phactr1 |
phosphatase and actin regulator 1 |
13978 |
0.24 |
| chr8_61588683_61589079 | 0.19 |
Palld |
palladin, cytoskeletal associated protein |
2258 |
0.41 |
| chr10_81414361_81414714 | 0.19 |
Mir1191b |
microRNA 1191b |
1760 |
0.13 |
| chr2_177468355_177468573 | 0.19 |
Zfp970 |
zinc finger protein 970 |
3618 |
0.19 |
| chr12_25177822_25178120 | 0.19 |
Gm47705 |
predicted gene, 47705 |
3797 |
0.22 |
| chr12_99345873_99346024 | 0.19 |
3300002A11Rik |
RIKEN cDNA 3300002A11 gene |
450 |
0.75 |
| chr19_32974465_32974785 | 0.19 |
Gm36860 |
predicted gene, 36860 |
5438 |
0.29 |
| chr8_121568635_121568786 | 0.19 |
Fbxo31 |
F-box protein 31 |
8747 |
0.11 |
| chr18_66076747_66077057 | 0.19 |
Ccbe1 |
collagen and calcium binding EGF domains 1 |
499 |
0.78 |
| chr10_83310985_83311139 | 0.19 |
Slc41a2 |
solute carrier family 41, member 2 |
6687 |
0.19 |
| chr12_104100793_104100977 | 0.19 |
Serpina5 |
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
228 |
0.86 |
| chr1_51272730_51272882 | 0.18 |
Cavin2 |
caveolae associated 2 |
16320 |
0.19 |
| chr10_67051361_67051540 | 0.18 |
Reep3 |
receptor accessory protein 3 |
14128 |
0.18 |
| chr7_142714810_142714975 | 0.18 |
Ins2 |
insulin II |
15382 |
0.13 |
| chr8_46522524_46522828 | 0.18 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
4110 |
0.19 |
| chr10_127884248_127884860 | 0.18 |
Rdh7 |
retinol dehydrogenase 7 |
3776 |
0.12 |
| chr4_125420233_125420436 | 0.18 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
70366 |
0.1 |
| chr6_16668124_16668275 | 0.18 |
Gm36669 |
predicted gene, 36669 |
109325 |
0.07 |
| chr7_63899068_63899509 | 0.18 |
Gm27252 |
predicted gene 27252 |
1314 |
0.37 |
| chr5_90891820_90892079 | 0.18 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
706 |
0.49 |
| chr16_41202636_41202787 | 0.18 |
Gm26381 |
predicted gene, 26381 |
15034 |
0.24 |
| chr8_93063870_93064038 | 0.18 |
Ces1a |
carboxylesterase 1A |
15762 |
0.14 |
| chr12_30410767_30410918 | 0.18 |
Sntg2 |
syntrophin, gamma 2 |
37467 |
0.14 |
| chr6_114887034_114887225 | 0.18 |
Vgll4 |
vestigial like family member 4 |
12022 |
0.2 |
| chr14_41159825_41160404 | 0.18 |
Mbl1 |
mannose-binding lectin (protein A) 1 |
8613 |
0.11 |
| chr5_90588842_90588993 | 0.18 |
Gm42109 |
predicted gene, 42109 |
140 |
0.94 |
| chr1_67185796_67186060 | 0.18 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
62902 |
0.11 |
| chr14_8246841_8246992 | 0.18 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
486 |
0.81 |
| chr3_57296881_57297058 | 0.18 |
Tm4sf1 |
transmembrane 4 superfamily member 1 |
1844 |
0.35 |
| chr11_100931218_100931481 | 0.18 |
Stat3 |
signal transducer and activator of transcription 3 |
8031 |
0.14 |
| chr9_65191764_65191933 | 0.18 |
Parp16 |
poly (ADP-ribose) polymerase family, member 16 |
3505 |
0.15 |
| chr4_6913436_6913609 | 0.18 |
Tox |
thymocyte selection-associated high mobility group box |
76961 |
0.11 |
| chr5_98029045_98029196 | 0.18 |
Antxr2 |
anthrax toxin receptor 2 |
1842 |
0.27 |
| chr19_38283394_38283545 | 0.18 |
Gm50141 |
predicted gene, 50141 |
4482 |
0.18 |
| chr6_149138694_149138910 | 0.18 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
1047 |
0.41 |
| chr4_120073028_120073179 | 0.18 |
AL607142.1 |
novel protein |
8107 |
0.23 |
| chr10_12278450_12278601 | 0.17 |
Gm48723 |
predicted gene, 48723 |
46136 |
0.14 |
| chr11_61355295_61355921 | 0.17 |
Slc47a1 |
solute carrier family 47, member 1 |
10044 |
0.13 |
| chr6_51984231_51984382 | 0.17 |
Skap2 |
src family associated phosphoprotein 2 |
28209 |
0.14 |
| chr2_91072228_91072550 | 0.17 |
Slc39a13 |
solute carrier family 39 (metal ion transporter), member 13 |
1972 |
0.19 |
| chr13_45851297_45851486 | 0.17 |
Atxn1 |
ataxin 1 |
20897 |
0.23 |
| chr17_32493567_32493723 | 0.17 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
1487 |
0.3 |
| chr7_98351265_98351558 | 0.17 |
Tsku |
tsukushi, small leucine rich proteoglycan |
8668 |
0.17 |
| chr8_89263116_89263276 | 0.17 |
Gm5356 |
predicted pseudogene 5356 |
75636 |
0.11 |
| chr2_69466274_69466471 | 0.17 |
Lrp2 |
low density lipoprotein receptor-related protein 2 |
42773 |
0.15 |
| chr8_45330798_45331088 | 0.17 |
Cyp4v3 |
cytochrome P450, family 4, subfamily v, polypeptide 3 |
2273 |
0.24 |
| chr11_5524693_5525093 | 0.17 |
Xbp1 |
X-box binding protein 1 |
2970 |
0.18 |
| chr14_19987117_19987272 | 0.17 |
Gng2 |
guanine nucleotide binding protein (G protein), gamma 2 |
9567 |
0.21 |
| chr8_93193069_93193250 | 0.17 |
Gm45909 |
predicted gene 45909 |
1801 |
0.25 |
| chr7_121907300_121907714 | 0.17 |
Scnn1b |
sodium channel, nonvoltage-gated 1 beta |
8279 |
0.18 |
| chr2_167727310_167727515 | 0.17 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
36231 |
0.09 |
| chr13_31477017_31477445 | 0.17 |
Gm23351 |
predicted gene, 23351 |
49976 |
0.1 |
| chr4_97809720_97810073 | 0.17 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
31818 |
0.18 |
| chr13_67306829_67307011 | 0.17 |
Zfp457 |
zinc finger protein 457 |
435 |
0.64 |
| chr2_155235951_155236129 | 0.17 |
Dynlrb1 |
dynein light chain roadblock-type 1 |
493 |
0.77 |
| chr6_141994065_141994238 | 0.17 |
Gm6614 |
predicted gene 6614 |
14594 |
0.21 |
| chr4_140856184_140856335 | 0.17 |
Padi1 |
peptidyl arginine deiminase, type I |
10481 |
0.12 |
| chr7_98349652_98349832 | 0.17 |
Tsku |
tsukushi, small leucine rich proteoglycan |
10337 |
0.17 |
| chr5_150919964_150920115 | 0.17 |
Kl |
klotho |
32568 |
0.15 |
| chr4_80947580_80947746 | 0.17 |
Gm27452 |
predicted gene, 27452 |
19781 |
0.23 |
| chr15_88854629_88854809 | 0.17 |
Pim3 |
proviral integration site 3 |
7467 |
0.13 |
| chr3_85251802_85252139 | 0.17 |
Gm38313 |
predicted gene, 38313 |
55523 |
0.13 |
| chr14_121720038_121720281 | 0.17 |
Dock9 |
dedicator of cytokinesis 9 |
18702 |
0.23 |
| chr4_59002789_59002940 | 0.17 |
Dnajc25 |
DnaJ heat shock protein family (Hsp40) member C25 |
308 |
0.51 |
| chr3_49778482_49778633 | 0.17 |
Gm37550 |
predicted gene, 37550 |
2330 |
0.33 |
| chr2_131345745_131345918 | 0.16 |
Rnf24 |
ring finger protein 24 |
7031 |
0.14 |
| chr6_141432376_141432556 | 0.16 |
Gm43958 |
predicted gene, 43958 |
9894 |
0.26 |
| chr5_66337632_66338067 | 0.16 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
167 |
0.93 |
| chr6_17502652_17502835 | 0.16 |
Met |
met proto-oncogene |
11502 |
0.23 |
| chr2_132859056_132859242 | 0.16 |
Crls1 |
cardiolipin synthase 1 |
11411 |
0.13 |
| chr16_85172743_85172897 | 0.16 |
App |
amyloid beta (A4) precursor protein |
884 |
0.58 |
| chr8_3049184_3049482 | 0.16 |
Gm44634 |
predicted gene 44634 |
6961 |
0.2 |
| chr4_117131790_117132241 | 0.16 |
Plk3 |
polo like kinase 3 |
1784 |
0.13 |
| chr7_128629489_128629745 | 0.16 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
18257 |
0.11 |
| chr4_76942367_76942518 | 0.16 |
Gm11246 |
predicted gene 11246 |
15131 |
0.21 |
| chrX_169906579_169906730 | 0.16 |
Mid1 |
midline 1 |
20297 |
0.21 |
| chr10_99604268_99604433 | 0.16 |
Gm20110 |
predicted gene, 20110 |
4821 |
0.21 |
| chr19_40180011_40180165 | 0.16 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
7198 |
0.16 |
| chr17_64203273_64203432 | 0.16 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
109679 |
0.07 |
| chr3_79543354_79543505 | 0.16 |
Gm3513 |
predicted gene 3513 |
8764 |
0.14 |
| chr13_31506338_31507020 | 0.16 |
Foxq1 |
forkhead box Q1 |
49455 |
0.1 |
| chr11_103130580_103130749 | 0.16 |
Hexim2 |
hexamethylene bis-acetamide inducible 2 |
1765 |
0.23 |
| chr12_12271220_12271371 | 0.16 |
Fam49a |
family with sequence similarity 49, member A |
9106 |
0.29 |
| chr9_65291786_65291937 | 0.16 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
2399 |
0.16 |
| chr13_44430370_44430571 | 0.16 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
9242 |
0.15 |
| chr11_105235813_105236112 | 0.16 |
Tlk2 |
tousled-like kinase 2 (Arabidopsis) |
10796 |
0.19 |
| chr6_48742053_48742218 | 0.16 |
Gimap1 |
GTPase, IMAP family member 1 |
47 |
0.88 |
| chr2_90983757_90983908 | 0.16 |
Celf1 |
CUGBP, Elav-like family member 1 |
3788 |
0.15 |
| chr15_41832966_41833117 | 0.16 |
Oxr1 |
oxidation resistance 1 |
2048 |
0.35 |
| chr4_139336597_139336796 | 0.16 |
AL807811.1 |
aldo-keto reductase family 7 member A2 (AKR7A2) pseudogene |
484 |
0.54 |
| chr5_92149399_92149550 | 0.16 |
Gm15710 |
predicted gene 15710 |
8545 |
0.12 |
| chr1_188729295_188729459 | 0.16 |
Gm25269 |
predicted gene, 25269 |
31099 |
0.23 |
| chr9_47348407_47348558 | 0.16 |
Gm31816 |
predicted gene, 31816 |
14268 |
0.26 |
| chr16_31933221_31933374 | 0.16 |
Gm49731 |
predicted gene, 49731 |
350 |
0.56 |
| chr7_65337727_65337920 | 0.16 |
Tjp1 |
tight junction protein 1 |
4974 |
0.23 |
| chr9_119198752_119198924 | 0.16 |
Slc22a14 |
solute carrier family 22 (organic cation transporter), member 14 |
8431 |
0.12 |
| chr15_41948299_41948658 | 0.16 |
Gm24751 |
predicted gene, 24751 |
519 |
0.8 |
| chr10_20590619_20590770 | 0.16 |
Gm17230 |
predicted gene 17230 |
34941 |
0.16 |
| chr7_16792284_16792639 | 0.16 |
Slc1a5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
566 |
0.62 |
| chr5_65491933_65492390 | 0.16 |
Gm43552 |
predicted gene 43552 |
672 |
0.31 |
| chr10_107310865_107311060 | 0.16 |
Lin7a |
lin-7 homolog A (C. elegans) |
38568 |
0.18 |
| chr1_175664518_175664669 | 0.16 |
Chml |
choroideremia-like |
23760 |
0.13 |
| chr5_65600569_65600720 | 0.16 |
Ube2k |
ubiquitin-conjugating enzyme E2K |
9048 |
0.09 |
| chr2_5839502_5839908 | 0.16 |
Cdc123 |
cell division cycle 123 |
5146 |
0.16 |
| chr15_25187875_25188048 | 0.16 |
5830426I08Rik |
RIKEN cDNA 5830426I08 gene |
37893 |
0.13 |
| chr10_124590350_124590536 | 0.15 |
4930503E24Rik |
RIKEN cDNA 4930503E24 gene |
67005 |
0.14 |
| chr19_37177758_37177920 | 0.15 |
Cpeb3 |
cytoplasmic polyadenylation element binding protein 3 |
178 |
0.9 |
| chr16_66041495_66041684 | 0.15 |
Gm49635 |
predicted gene, 49635 |
94483 |
0.09 |
| chr4_71658237_71658395 | 0.15 |
Gm11231 |
predicted gene 11231 |
19697 |
0.27 |
| chr7_115859114_115859300 | 0.15 |
Sox6 |
SRY (sex determining region Y)-box 6 |
645 |
0.82 |
| chr16_91678296_91678553 | 0.15 |
Donson |
downstream neighbor of SON |
2886 |
0.15 |
| chr8_24456652_24456803 | 0.15 |
Gm44620 |
predicted gene 44620 |
9630 |
0.15 |
| chr6_146945287_146945499 | 0.15 |
1700034J05Rik |
RIKEN cDNA 1700034J05 gene |
9021 |
0.16 |
| chr4_86661560_86661711 | 0.15 |
Plin2 |
perilipin 2 |
2487 |
0.26 |
| chr7_131434431_131434598 | 0.15 |
Acadsb |
acyl-Coenzyme A dehydrogenase, short/branched chain |
2513 |
0.14 |
| chr7_81577348_81577499 | 0.15 |
Whamm |
WAS protein homolog associated with actin, golgi membranes and microtubules |
5479 |
0.11 |
| chr2_155056753_155056913 | 0.15 |
a |
nonagouti |
9218 |
0.14 |
| chr5_9042843_9042994 | 0.15 |
Gm40264 |
predicted gene, 40264 |
7794 |
0.15 |
| chr3_22168181_22168357 | 0.15 |
Tbl1xr1 |
transducin (beta)-like 1X-linked receptor 1 |
19327 |
0.21 |
| chr9_58554849_58555000 | 0.15 |
Gm10657 |
predicted gene 10657 |
125 |
0.85 |
| chr11_77417847_77418056 | 0.15 |
Ssh2 |
slingshot protein phosphatase 2 |
26514 |
0.13 |
| chr14_30929998_30930386 | 0.15 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
6432 |
0.11 |
| chr5_20858888_20859039 | 0.15 |
Phtf2 |
putative homeodomain transcription factor 2 |
3772 |
0.22 |
| chr1_171341329_171341480 | 0.15 |
Nit1 |
nitrilase 1 |
3063 |
0.08 |
| chr6_118478873_118479106 | 0.15 |
Zfp9 |
zinc finger protein 9 |
331 |
0.85 |
| chr19_37456339_37456518 | 0.15 |
Gm23026 |
predicted gene, 23026 |
7318 |
0.13 |
| chr2_3714011_3714368 | 0.15 |
Fam107b |
family with sequence similarity 107, member B |
711 |
0.65 |
| chr1_21240946_21241097 | 0.15 |
Gsta3 |
glutathione S-transferase, alpha 3 |
392 |
0.76 |
| chr14_61680870_61681697 | 0.15 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr1_67202559_67202731 | 0.15 |
Gm15668 |
predicted gene 15668 |
46555 |
0.14 |
| chr13_92050387_92050727 | 0.15 |
Rasgrf2 |
RAS protein-specific guanine nucleotide-releasing factor 2 |
19653 |
0.26 |
| chr16_91785546_91785722 | 0.15 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
1529 |
0.38 |
| chr15_85803009_85803377 | 0.15 |
Cdpf1 |
cysteine rich, DPF motif domain containing 1 |
7897 |
0.13 |
| chr13_9016873_9017115 | 0.15 |
Gtpbp4 |
GTP binding protein 4 |
20911 |
0.1 |
| chr14_70509802_70509953 | 0.15 |
Bmp1 |
bone morphogenetic protein 1 |
1091 |
0.34 |
| chr8_122696911_122697102 | 0.15 |
Gm10612 |
predicted gene 10612 |
854 |
0.41 |
| chr17_22739370_22739536 | 0.15 |
Gm5493 |
predicted gene 5493 |
492 |
0.74 |
| chr19_12708659_12708815 | 0.15 |
Gm15962 |
predicted gene 15962 |
12466 |
0.09 |
| chr16_49531828_49532031 | 0.15 |
Gm6931 |
predicted gene 6931 |
107123 |
0.07 |
| chr4_119551551_119551702 | 0.15 |
Gm12959 |
predicted gene 12959 |
2432 |
0.16 |
| chr2_31503965_31504231 | 0.15 |
Ass1 |
argininosuccinate synthetase 1 |
3372 |
0.23 |
| chr14_65718336_65718507 | 0.14 |
Scara5 |
scavenger receptor class A, member 5 |
41944 |
0.15 |
| chr2_166032901_166033054 | 0.14 |
Ncoa3 |
nuclear receptor coactivator 3 |
14941 |
0.15 |
| chr18_7894073_7894247 | 0.14 |
Wac |
WW domain containing adaptor with coiled-coil |
11315 |
0.17 |
| chr16_30053012_30053514 | 0.14 |
Hes1 |
hes family bHLH transcription factor 1 |
11121 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.2 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.1 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.2 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.2 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:2000347 | positive regulation of hepatocyte proliferation(GO:2000347) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.0 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.0 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid metabolic process(GO:0009698) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.0 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.0 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.0 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.0 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.0 | GO:0045293 | mRNA editing complex(GO:0045293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.6 | GO:0080030 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) 3,4-dihydrocoumarin hydrolase activity(GO:0018733) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.0 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |