Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
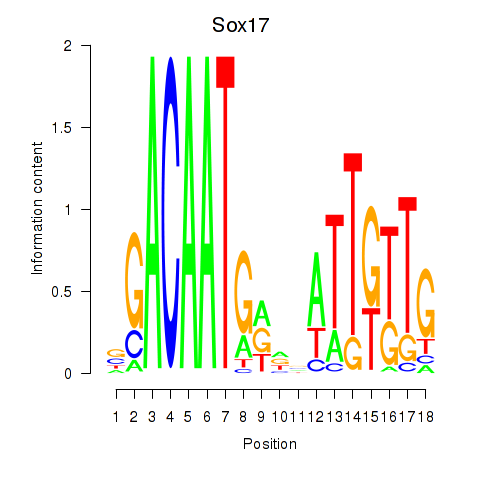
Results for Sox17
Z-value: 1.53
Transcription factors associated with Sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox17
|
ENSMUSG00000025902.7 | SRY (sex determining region Y)-box 17 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_4479698_4479869 | Sox17 | 13821 | 0.141728 | -0.71 | 1.1e-01 | Click! |
Activity of the Sox17 motif across conditions
Conditions sorted by the z-value of the Sox17 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
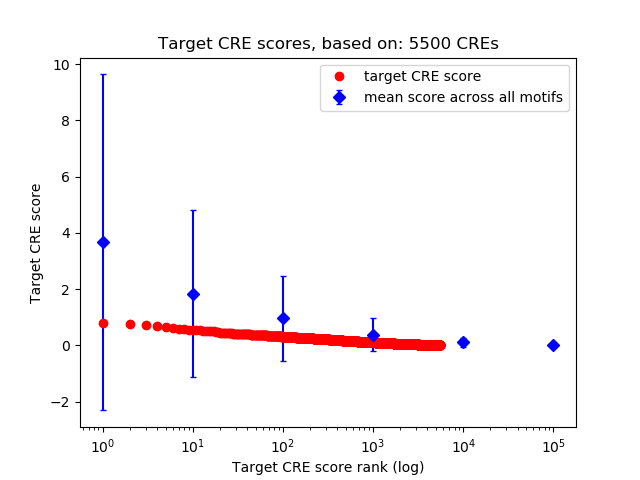
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_62909790_62909949 | 0.78 |
Fbp1 |
fructose bisphosphatase 1 |
21587 |
0.13 |
| chr3_138289329_138289480 | 0.76 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
11753 |
0.11 |
| chr12_69941545_69941731 | 0.73 |
Atl1 |
atlastin GTPase 1 |
4539 |
0.13 |
| chr17_62789514_62789722 | 0.68 |
Efna5 |
ephrin A5 |
91526 |
0.1 |
| chr1_87339717_87339868 | 0.66 |
Gm22549 |
predicted gene, 22549 |
1769 |
0.26 |
| chr16_43168807_43168974 | 0.61 |
Gm15712 |
predicted gene 15712 |
15683 |
0.21 |
| chr3_94339066_94339223 | 0.57 |
Them5 |
thioesterase superfamily member 5 |
2955 |
0.11 |
| chr10_83310985_83311139 | 0.57 |
Slc41a2 |
solute carrier family 41, member 2 |
6687 |
0.19 |
| chr16_43276603_43276913 | 0.56 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
29474 |
0.15 |
| chr19_26823384_26823535 | 0.55 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
448 |
0.84 |
| chr5_67019971_67020122 | 0.55 |
Gm42713 |
predicted gene 42713 |
19330 |
0.12 |
| chr12_57540843_57541103 | 0.54 |
Foxa1 |
forkhead box A1 |
5148 |
0.17 |
| chr15_3506426_3506577 | 0.52 |
Ghr |
growth hormone receptor |
34857 |
0.21 |
| chr7_80625574_80625744 | 0.52 |
Crtc3 |
CREB regulated transcription coactivator 3 |
3967 |
0.19 |
| chr12_45065771_45065940 | 0.51 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
8257 |
0.21 |
| chr1_21260733_21261242 | 0.51 |
Gsta3 |
glutathione S-transferase, alpha 3 |
7466 |
0.11 |
| chr12_69941731_69941931 | 0.51 |
Atl1 |
atlastin GTPase 1 |
4346 |
0.13 |
| chr4_53039519_53039670 | 0.48 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
1332 |
0.41 |
| chr3_97649987_97650151 | 0.46 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
8124 |
0.13 |
| chr9_9565147_9565311 | 0.45 |
Gm47334 |
predicted gene, 47334 |
1142 |
0.57 |
| chr15_3458621_3458946 | 0.45 |
Ghr |
growth hormone receptor |
12861 |
0.28 |
| chr11_68865350_68865528 | 0.44 |
Ndel1 |
nudE neurodevelopment protein 1 like 1 |
446 |
0.74 |
| chr6_90469663_90469814 | 0.44 |
Klf15 |
Kruppel-like factor 15 |
2620 |
0.15 |
| chr12_40230630_40230800 | 0.44 |
Gm47954 |
predicted gene, 47954 |
4933 |
0.12 |
| chr8_127097367_127097518 | 0.43 |
Pard3 |
par-3 family cell polarity regulator |
24202 |
0.18 |
| chr1_60609032_60609279 | 0.43 |
Gm23762 |
predicted gene, 23762 |
4648 |
0.15 |
| chr9_112027249_112027400 | 0.42 |
Mir128-2 |
microRNA 128-2 |
91387 |
0.08 |
| chr14_10751127_10751283 | 0.42 |
Fhitos |
fragile histidine triad gene, opposite strand |
12652 |
0.25 |
| chr1_67201803_67201976 | 0.42 |
Gm15668 |
predicted gene 15668 |
47311 |
0.14 |
| chr13_75543303_75543454 | 0.42 |
Gm3926 |
predicted gene 3926 |
19030 |
0.15 |
| chr1_39425203_39425354 | 0.42 |
Gm37265 |
predicted gene, 37265 |
5346 |
0.19 |
| chr16_32896347_32896498 | 0.41 |
Fyttd1 |
forty-two-three domain containing 1 |
1331 |
0.34 |
| chr7_125256750_125256907 | 0.41 |
Gm21957 |
predicted gene, 21957 |
36942 |
0.16 |
| chr15_4836304_4836788 | 0.41 |
Gm49074 |
predicted gene, 49074 |
10841 |
0.18 |
| chr4_148617242_148617393 | 0.41 |
Tardbp |
TAR DNA binding protein |
337 |
0.8 |
| chr12_8796924_8797075 | 0.40 |
Sdc1 |
syndecan 1 |
25196 |
0.16 |
| chr7_132924974_132925182 | 0.40 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
6025 |
0.15 |
| chr5_90891820_90892079 | 0.39 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
706 |
0.49 |
| chr10_20024835_20025010 | 0.39 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
73867 |
0.09 |
| chr13_46676987_46677175 | 0.39 |
Nup153 |
nucleoporin 153 |
4871 |
0.18 |
| chr2_11712487_11713017 | 0.39 |
Il15ra |
interleukin 15 receptor, alpha chain |
5575 |
0.16 |
| chr14_14789749_14789914 | 0.39 |
Slc4a7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
3904 |
0.26 |
| chr13_36533643_36534408 | 0.39 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
3072 |
0.22 |
| chr17_64765537_64765696 | 0.38 |
Dreh |
down-regulated in hepatocellular carcinoma |
1495 |
0.41 |
| chr19_29520625_29520778 | 0.38 |
A930007I19Rik |
RIKEN cDNA A930007I19 gene |
269 |
0.89 |
| chr7_30943687_30943953 | 0.38 |
Hamp |
hepcidin antimicrobial peptide |
212 |
0.82 |
| chr9_57255390_57255612 | 0.38 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
6008 |
0.17 |
| chr13_112500358_112500528 | 0.38 |
Il6st |
interleukin 6 signal transducer |
25397 |
0.14 |
| chr2_181461420_181461583 | 0.38 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
2075 |
0.19 |
| chr14_117447795_117448102 | 0.38 |
Mir6239 |
microRNA 6239 |
505899 |
0.0 |
| chr4_150393280_150393601 | 0.38 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
12954 |
0.22 |
| chr8_122985769_122985939 | 0.37 |
Gm24445 |
predicted gene, 24445 |
2911 |
0.19 |
| chr9_55541381_55541949 | 0.37 |
Isl2 |
insulin related protein 2 (islet 2) |
458 |
0.61 |
| chr18_36656725_36657021 | 0.37 |
Ankhd1 |
ankyrin repeat and KH domain containing 1 |
1363 |
0.23 |
| chr2_167727310_167727515 | 0.37 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
36231 |
0.09 |
| chr14_34675574_34675754 | 0.37 |
Wapl |
WAPL cohesin release factor |
1491 |
0.26 |
| chr9_55253735_55254087 | 0.37 |
Nrg4 |
neuregulin 4 |
11450 |
0.18 |
| chr4_142337823_142337991 | 0.37 |
Gm13052 |
predicted gene 13052 |
8568 |
0.28 |
| chr9_94083843_94084083 | 0.37 |
Gm5369 |
predicted gene 5369 |
55499 |
0.17 |
| chr11_31722665_31722816 | 0.36 |
Gm38061 |
predicted gene, 38061 |
24092 |
0.2 |
| chr19_12697110_12697264 | 0.36 |
Keg1 |
kidney expressed gene 1 |
1373 |
0.26 |
| chr15_7135727_7135925 | 0.36 |
Lifr |
LIF receptor alpha |
4716 |
0.31 |
| chr8_46511132_46511387 | 0.36 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
15527 |
0.15 |
| chr10_31497285_31497485 | 0.36 |
Gm47687 |
predicted gene, 47687 |
45303 |
0.1 |
| chr3_141502934_141503090 | 0.36 |
Unc5c |
unc-5 netrin receptor C |
37341 |
0.18 |
| chr11_111998446_111998808 | 0.35 |
Gm11679 |
predicted gene 11679 |
44951 |
0.19 |
| chr6_48542267_48542478 | 0.35 |
Atp6v0e2 |
ATPase, H+ transporting, lysosomal V0 subunit E2 |
4707 |
0.08 |
| chr2_131345745_131345918 | 0.34 |
Rnf24 |
ring finger protein 24 |
7031 |
0.14 |
| chr7_119670801_119670968 | 0.34 |
Acsm4 |
acyl-CoA synthetase medium-chain family member 4 |
19142 |
0.1 |
| chr8_12897974_12898151 | 0.34 |
Gm15353 |
predicted gene 15353 |
14336 |
0.11 |
| chr4_83418470_83418621 | 0.34 |
Snapc3 |
small nuclear RNA activating complex, polypeptide 3 |
796 |
0.58 |
| chr2_49177399_49177550 | 0.34 |
Gm13510 |
predicted gene 13510 |
7544 |
0.27 |
| chr8_73938665_73938983 | 0.34 |
Gm7948 |
predicted gene 7948 |
67131 |
0.14 |
| chr8_42183376_42183557 | 0.33 |
Gm6180 |
predicted pseudogene 6180 |
63445 |
0.14 |
| chr19_55260521_55260708 | 0.33 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
7245 |
0.18 |
| chr10_20010162_20010316 | 0.33 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
59184 |
0.12 |
| chr1_3586531_3586701 | 0.33 |
Gm38148 |
predicted gene, 38148 |
9287 |
0.19 |
| chr8_70207953_70208123 | 0.33 |
Slc25a42 |
solute carrier family 25, member 42 |
4218 |
0.12 |
| chr11_44518397_44518764 | 0.33 |
Rnf145 |
ring finger protein 145 |
384 |
0.85 |
| chr3_138290241_138291213 | 0.33 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
13076 |
0.11 |
| chr16_44137078_44137229 | 0.33 |
Atp6v1a |
ATPase, H+ transporting, lysosomal V1 subunit A |
1827 |
0.3 |
| chr15_4678897_4679270 | 0.33 |
C6 |
complement component 6 |
48092 |
0.17 |
| chr5_123091308_123091459 | 0.33 |
Tmem120b |
transmembrane protein 120B |
8518 |
0.09 |
| chr9_53192914_53193065 | 0.33 |
Gm47986 |
predicted gene, 47986 |
28801 |
0.18 |
| chr2_135855339_135855516 | 0.32 |
Plcb4 |
phospholipase C, beta 4 |
32267 |
0.19 |
| chr16_43323583_43323748 | 0.32 |
Gm15711 |
predicted gene 15711 |
11071 |
0.17 |
| chr8_114663169_114663337 | 0.32 |
Gm16117 |
predicted gene 16117 |
21139 |
0.19 |
| chr10_68102616_68102812 | 0.32 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
33912 |
0.17 |
| chr8_107311781_107311960 | 0.32 |
Nfat5 |
nuclear factor of activated T cells 5 |
18311 |
0.18 |
| chr3_84647383_84647687 | 0.32 |
Tmem154 |
transmembrane protein 154 |
18657 |
0.2 |
| chr5_17280342_17280505 | 0.32 |
Gm6673 |
predicted gene 6673 |
52489 |
0.15 |
| chr7_28959292_28959443 | 0.32 |
Actn4 |
actinin alpha 4 |
2856 |
0.14 |
| chr2_27710029_27710632 | 0.32 |
Rxra |
retinoid X receptor alpha |
253 |
0.95 |
| chr12_80837168_80837319 | 0.32 |
Susd6 |
sushi domain containing 6 |
31465 |
0.12 |
| chr6_108281132_108281339 | 0.31 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
28277 |
0.22 |
| chr10_75892623_75892818 | 0.31 |
Derl3 |
Der1-like domain family, member 3 |
693 |
0.47 |
| chr17_34852319_34852771 | 0.31 |
Nelfe |
negative elongation factor complex member E, Rdbp |
63 |
0.89 |
| chr9_44946661_44946812 | 0.31 |
Gm19121 |
predicted gene, 19121 |
8979 |
0.08 |
| chr6_101326488_101326738 | 0.31 |
Gm43953 |
predicted gene, 43953 |
1592 |
0.32 |
| chr10_24923327_24923737 | 0.31 |
Arg1 |
arginase, liver |
3905 |
0.16 |
| chr9_122149132_122149346 | 0.30 |
Gm47121 |
predicted gene, 47121 |
6790 |
0.13 |
| chr4_124431592_124431891 | 0.30 |
1700057H15Rik |
RIKEN cDNA 1700057H15 gene |
12883 |
0.21 |
| chr11_11845100_11845416 | 0.30 |
Ddc |
dopa decarboxylase |
8978 |
0.18 |
| chr4_48909780_48909970 | 0.30 |
Gm12436 |
predicted gene 12436 |
11282 |
0.2 |
| chr4_63267695_63267867 | 0.30 |
Mir455 |
microRNA 455 |
10930 |
0.16 |
| chr9_9173231_9173382 | 0.30 |
Gm16833 |
predicted gene, 16833 |
62982 |
0.13 |
| chr5_117364085_117364236 | 0.30 |
Wsb2 |
WD repeat and SOCS box-containing 2 |
638 |
0.56 |
| chr11_110249489_110249640 | 0.30 |
Abca6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
2212 |
0.39 |
| chr17_26253883_26254034 | 0.30 |
Luc7l |
Luc7-like |
736 |
0.39 |
| chr19_43811775_43811943 | 0.30 |
Abcc2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
1337 |
0.38 |
| chr7_123642615_123642780 | 0.30 |
Zkscan2 |
zinc finger with KRAB and SCAN domains 2 |
142248 |
0.04 |
| chr3_87001052_87001203 | 0.30 |
Cd1d1 |
CD1d1 antigen |
1686 |
0.28 |
| chr6_5176021_5176208 | 0.30 |
Pon1 |
paraoxonase 1 |
17649 |
0.18 |
| chr1_130877275_130877426 | 0.30 |
Fcmr |
Fc fragment of IgM receptor |
808 |
0.52 |
| chr10_19946542_19946693 | 0.30 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
4438 |
0.24 |
| chr4_96662396_96662547 | 0.30 |
Cyp2j5 |
cytochrome P450, family 2, subfamily j, polypeptide 5 |
1683 |
0.46 |
| chr1_139780384_139780535 | 0.30 |
Gm4788 |
predicted gene 4788 |
709 |
0.69 |
| chr17_47976252_47976712 | 0.29 |
Gm14871 |
predicted gene 14871 |
27090 |
0.11 |
| chr11_7815566_7815774 | 0.29 |
Gm27393 |
predicted gene, 27393 |
70835 |
0.13 |
| chr6_28471887_28472038 | 0.29 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
3177 |
0.17 |
| chr9_72702491_72702671 | 0.29 |
Nedd4 |
neural precursor cell expressed, developmentally down-regulated 4 |
9142 |
0.1 |
| chr1_133650504_133650728 | 0.29 |
Zc3h11a |
zinc finger CCCH type containing 11A |
10665 |
0.13 |
| chr18_69393942_69394102 | 0.29 |
Tcf4 |
transcription factor 4 |
21875 |
0.25 |
| chr10_40446168_40446390 | 0.29 |
Cdk19os |
cyclin-dependent kinase 19, opposite strand |
96176 |
0.05 |
| chr5_148633016_148633167 | 0.29 |
Gm29815 |
predicted gene, 29815 |
19447 |
0.18 |
| chr5_17963645_17963839 | 0.29 |
Gnat3 |
guanine nucleotide binding protein, alpha transducing 3 |
1193 |
0.63 |
| chr16_49857039_49857216 | 0.29 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
1340 |
0.56 |
| chr19_4772512_4772663 | 0.29 |
Gm37206 |
predicted gene, 37206 |
7416 |
0.09 |
| chr8_36280569_36280720 | 0.28 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
31128 |
0.17 |
| chr8_5030377_5030626 | 0.28 |
n-R5s93 |
nuclear encoded rRNA 5S 93 |
38876 |
0.14 |
| chr5_8164154_8164307 | 0.28 |
Gm21759 |
predicted gene, 21759 |
15406 |
0.17 |
| chr4_53222171_53222454 | 0.28 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
4455 |
0.21 |
| chr18_32559226_32559588 | 0.28 |
Gypc |
glycophorin C |
573 |
0.78 |
| chr6_147664470_147664629 | 0.28 |
1700049E15Rik |
RIKEN cDNA 1700049E15 gene |
25692 |
0.21 |
| chr11_26684255_26684421 | 0.28 |
Gm12071 |
predicted gene 12071 |
28567 |
0.2 |
| chr14_17914680_17914831 | 0.28 |
Thrb |
thyroid hormone receptor beta |
48066 |
0.16 |
| chr9_68827763_68827979 | 0.28 |
Rora |
RAR-related orphan receptor alpha |
172568 |
0.03 |
| chr13_109635481_109635828 | 0.28 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
2874 |
0.42 |
| chr16_95141907_95142069 | 0.28 |
Gm49642 |
predicted gene, 49642 |
29522 |
0.18 |
| chr16_59413660_59413813 | 0.28 |
Gabrr3 |
gamma-aminobutyric acid (GABA) receptor, rho 3 |
6404 |
0.16 |
| chr19_55719676_55719857 | 0.28 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
22054 |
0.26 |
| chr1_69801581_69801779 | 0.28 |
Spag16 |
sperm associated antigen 16 |
25290 |
0.2 |
| chr1_151689614_151690051 | 0.27 |
Fam129a |
family with sequence similarity 129, member A |
12665 |
0.21 |
| chr9_71905527_71905697 | 0.27 |
Gm18821 |
predicted gene, 18821 |
3231 |
0.15 |
| chr4_154143077_154143228 | 0.27 |
Wrap73 |
WD repeat containing, antisense to Trp73 |
743 |
0.5 |
| chr12_86516484_86516661 | 0.27 |
Esrrb |
estrogen related receptor, beta |
12163 |
0.24 |
| chr2_167843295_167843525 | 0.27 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
9764 |
0.16 |
| chr7_30962949_30963144 | 0.27 |
Gm4673 |
predicted gene 4673 |
5637 |
0.07 |
| chr10_68126588_68126805 | 0.27 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
9930 |
0.25 |
| chr3_98741469_98742014 | 0.27 |
Hsd3b3 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
11724 |
0.13 |
| chr16_91618278_91618429 | 0.27 |
Dnajc28 |
DnaJ heat shock protein family (Hsp40) member C28 |
638 |
0.54 |
| chr6_72511397_72511595 | 0.27 |
Sh2d6 |
SH2 domain containing 6 |
8519 |
0.13 |
| chr13_35469616_35469777 | 0.27 |
Gm48704 |
predicted gene, 48704 |
74206 |
0.09 |
| chr13_9073792_9074124 | 0.27 |
Gm36264 |
predicted gene, 36264 |
2493 |
0.23 |
| chr1_23237656_23237879 | 0.27 |
Gm29506 |
predicted gene 29506 |
2094 |
0.23 |
| chr12_40574426_40574700 | 0.27 |
Dock4 |
dedicator of cytokinesis 4 |
128227 |
0.05 |
| chr5_65068578_65068729 | 0.27 |
Tmem156 |
transmembrane protein 156 |
22931 |
0.15 |
| chr17_56105602_56105761 | 0.27 |
Plin4 |
perilipin 4 |
4121 |
0.1 |
| chr1_16107096_16107247 | 0.27 |
Rdh10 |
retinol dehydrogenase 10 (all-trans) |
1397 |
0.36 |
| chr7_107699185_107699336 | 0.27 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
33949 |
0.12 |
| chr3_67085309_67085464 | 0.27 |
Gm22295 |
predicted gene, 22295 |
29428 |
0.2 |
| chr9_114857411_114857562 | 0.27 |
Cmtm8 |
CKLF-like MARVEL transmembrane domain containing 8 |
13330 |
0.16 |
| chr1_6511141_6511292 | 0.27 |
St18 |
suppression of tumorigenicity 18 |
23885 |
0.22 |
| chr2_163615957_163616108 | 0.26 |
Ttpal |
tocopherol (alpha) transfer protein-like |
4512 |
0.16 |
| chr16_45987509_45987734 | 0.26 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
22597 |
0.14 |
| chr4_43724876_43725040 | 0.26 |
Hrct1 |
histidine rich carboxyl terminus 1 |
2230 |
0.13 |
| chr3_87620282_87621117 | 0.26 |
Arhgef11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
1938 |
0.27 |
| chr16_38273526_38273677 | 0.26 |
Nr1i2 |
nuclear receptor subfamily 1, group I, member 2 |
21223 |
0.13 |
| chr5_138013172_138013323 | 0.26 |
Gm29127 |
predicted gene 29127 |
4172 |
0.1 |
| chr9_77723756_77723924 | 0.26 |
Gm47790 |
predicted gene, 47790 |
6439 |
0.15 |
| chr18_62095425_62095728 | 0.26 |
Gm41750 |
predicted gene, 41750 |
47042 |
0.14 |
| chr18_7644384_7644571 | 0.26 |
Mpp7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
17611 |
0.21 |
| chr15_10211684_10211835 | 0.26 |
Prlr |
prolactin receptor |
1706 |
0.5 |
| chr3_157730079_157730297 | 0.26 |
Gm33466 |
predicted gene, 33466 |
3780 |
0.29 |
| chr10_24227305_24227456 | 0.26 |
Moxd1 |
monooxygenase, DBH-like 1 |
3835 |
0.24 |
| chr7_13637528_13637679 | 0.26 |
Zbed4-ps1 |
zinc finger, BED type containing 4, pseudogene 1 |
1504 |
0.32 |
| chr8_93193292_93193577 | 0.26 |
Gm45909 |
predicted gene 45909 |
2076 |
0.23 |
| chr10_31827348_31827499 | 0.26 |
Gm47704 |
predicted gene, 47704 |
40489 |
0.2 |
| chr12_78209771_78209951 | 0.26 |
Gm6657 |
predicted gene 6657 |
8895 |
0.15 |
| chr8_71676856_71677014 | 0.26 |
Jak3 |
Janus kinase 3 |
383 |
0.7 |
| chr16_31933221_31933374 | 0.26 |
Gm49731 |
predicted gene, 49731 |
350 |
0.56 |
| chr1_73935593_73935760 | 0.26 |
Tns1 |
tensin 1 |
4865 |
0.25 |
| chr10_120247499_120247740 | 0.26 |
Llph |
LLP homolog, long-term synaptic facilitation (Aplysia) |
20433 |
0.12 |
| chr1_190156909_190157060 | 0.26 |
Gm28172 |
predicted gene 28172 |
11686 |
0.18 |
| chr2_160362942_160363707 | 0.26 |
Mafb |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
3741 |
0.3 |
| chr9_106269146_106269548 | 0.26 |
Poc1a |
POC1 centriolar protein A |
11714 |
0.11 |
| chr7_99985802_99985987 | 0.26 |
Rnf169 |
ring finger protein 169 |
5446 |
0.14 |
| chr3_18140093_18140313 | 0.26 |
Gm23686 |
predicted gene, 23686 |
37422 |
0.16 |
| chr17_56104602_56104753 | 0.26 |
Hdgfl2 |
HDGF like 2 |
5115 |
0.09 |
| chr17_78915849_78916004 | 0.26 |
Cebpzos |
CCAAT/enhancer binding protein (C/EBP), zeta, opposite strand |
574 |
0.63 |
| chr4_150709744_150709942 | 0.25 |
Gm16079 |
predicted gene 16079 |
31051 |
0.16 |
| chr1_39594759_39594927 | 0.25 |
Gm5100 |
predicted gene 5100 |
3162 |
0.16 |
| chr13_3889050_3889201 | 0.25 |
Net1 |
neuroepithelial cell transforming gene 1 |
1300 |
0.32 |
| chr8_35212732_35212892 | 0.25 |
Gm34474 |
predicted gene, 34474 |
5826 |
0.16 |
| chr16_43394836_43395188 | 0.25 |
Gm15713 |
predicted gene 15713 |
25184 |
0.16 |
| chr10_67100073_67100416 | 0.25 |
Reep3 |
receptor accessory protein 3 |
3299 |
0.24 |
| chr5_150593024_150593258 | 0.25 |
N4bp2l1 |
NEDD4 binding protein 2-like 1 |
628 |
0.52 |
| chr3_35931210_35931361 | 0.25 |
Dcun1d1 |
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
173 |
0.9 |
| chrX_85828106_85828330 | 0.25 |
Gm14762 |
predicted gene 14762 |
1557 |
0.31 |
| chr10_77629279_77629435 | 0.25 |
Ube2g2 |
ubiquitin-conjugating enzyme E2G 2 |
6979 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.4 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.5 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.3 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.2 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.1 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.0 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.0 | 0.1 | GO:1901859 | negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.0 | 0.1 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0035935 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.0 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 | 0.0 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.0 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0015927 | trehalase activity(GO:0015927) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |