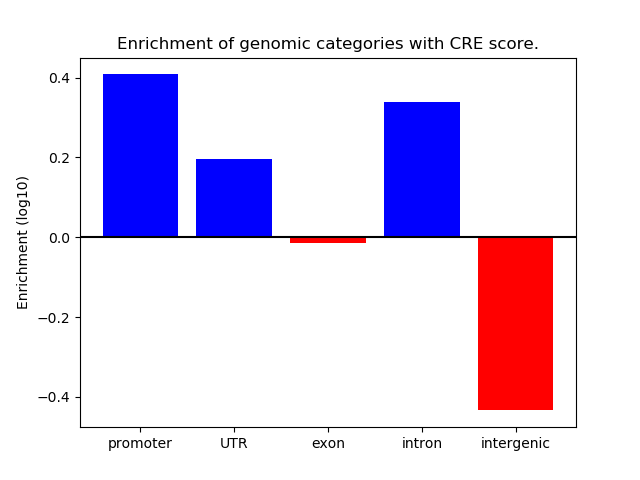
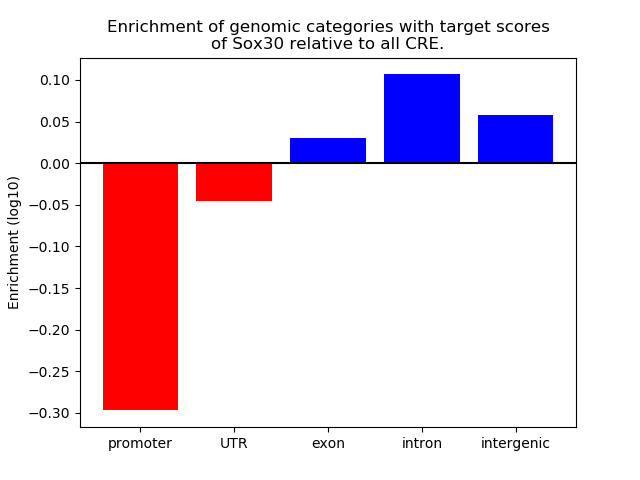
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
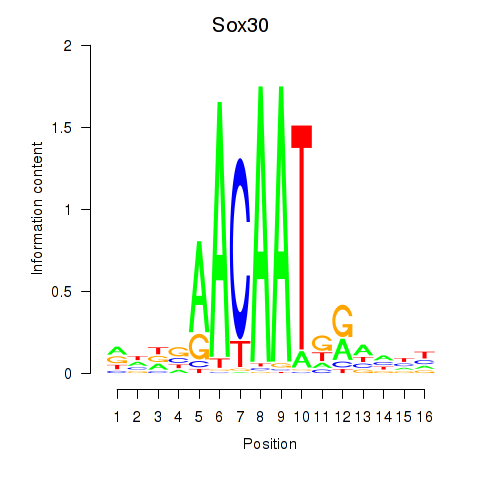
Results for Sox30
Z-value: 1.21
Transcription factors associated with Sox30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox30
|
ENSMUSG00000040489.5 | SRY (sex determining region Y)-box 30 |
Activity of the Sox30 motif across conditions
Conditions sorted by the z-value of the Sox30 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
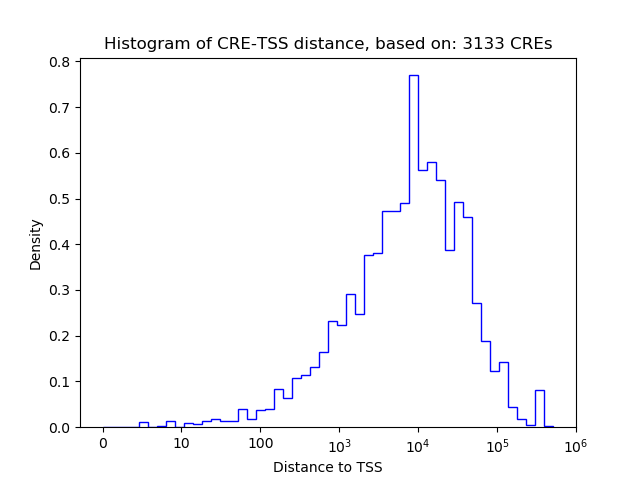
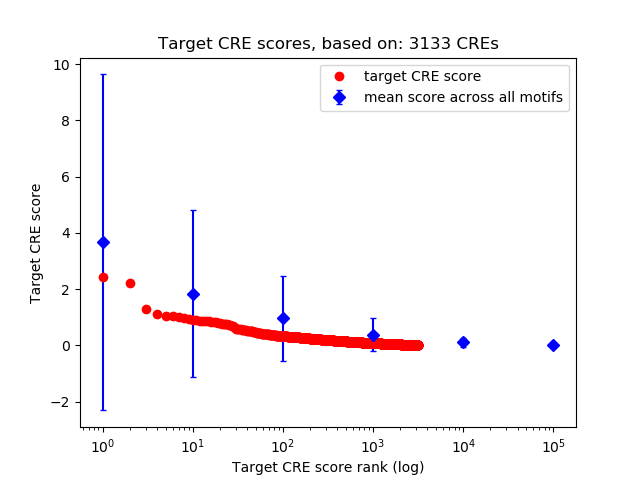
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_104346326_104346873 | 2.42 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
8113 |
0.12 |
| chr14_116442003_116442157 | 2.22 |
Gm38045 |
predicted gene, 38045 |
308135 |
0.01 |
| chr10_87920604_87920755 | 1.28 |
Tyms-ps |
thymidylate synthase, pseudogene |
46168 |
0.12 |
| chr12_40574426_40574700 | 1.11 |
Dock4 |
dedicator of cytokinesis 4 |
128227 |
0.05 |
| chr19_32992433_32992599 | 1.04 |
Gm36860 |
predicted gene, 36860 |
12453 |
0.25 |
| chr12_80010993_80011167 | 1.03 |
Gm8275 |
predicted gene 8275 |
31229 |
0.14 |
| chr13_24359929_24360183 | 0.99 |
Gm11342 |
predicted gene 11342 |
15894 |
0.12 |
| chr5_151018980_151019139 | 0.99 |
Gm8675 |
predicted gene 8675 |
9758 |
0.23 |
| chr2_72206740_72207276 | 0.95 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
8209 |
0.18 |
| chr4_65080374_65080537 | 0.92 |
Pappa |
pregnancy-associated plasma protein A |
43719 |
0.19 |
| chr5_125522928_125523080 | 0.90 |
Aacs |
acetoacetyl-CoA synthetase |
7761 |
0.16 |
| chr16_81366011_81366177 | 0.88 |
Gm49555 |
predicted gene, 49555 |
75932 |
0.11 |
| chr3_99188109_99188313 | 0.87 |
Gm18982 |
predicted gene, 18982 |
6930 |
0.13 |
| chr1_76641343_76641507 | 0.86 |
Mir6343 |
microRNA 6343 |
131865 |
0.05 |
| chr5_38117047_38117323 | 0.85 |
Stx18 |
syntaxin 18 |
3724 |
0.2 |
| chr11_16839622_16839885 | 0.85 |
Egfros |
epidermal growth factor receptor, opposite strand |
9051 |
0.21 |
| chr5_87349083_87349430 | 0.82 |
Gm42796 |
predicted gene 42796 |
6579 |
0.11 |
| chr1_41559831_41559986 | 0.82 |
Gm28634 |
predicted gene 28634 |
30365 |
0.26 |
| chr4_49550356_49550507 | 0.80 |
Aldob |
aldolase B, fructose-bisphosphate |
885 |
0.51 |
| chr18_56396921_56397088 | 0.79 |
Gramd3 |
GRAM domain containing 3 |
3333 |
0.25 |
| chr9_122865001_122865302 | 0.77 |
Zfp445 |
zinc finger protein 445 |
783 |
0.46 |
| chr19_33489637_33489788 | 0.76 |
Lipo5 |
lipase, member O5 |
16521 |
0.14 |
| chr17_63010676_63010827 | 0.75 |
Gm25348 |
predicted gene, 25348 |
86249 |
0.1 |
| chr2_58784544_58784695 | 0.75 |
Upp2 |
uridine phosphorylase 2 |
19294 |
0.19 |
| chr19_33480626_33480777 | 0.73 |
Lipo5 |
lipase, member O5 |
7510 |
0.16 |
| chr3_24378479_24378630 | 0.72 |
Gm7536 |
predicted gene 7536 |
45486 |
0.21 |
| chr1_71807937_71808302 | 0.68 |
Gm37217 |
predicted gene, 37217 |
38491 |
0.15 |
| chr1_67138759_67139000 | 0.68 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
15853 |
0.23 |
| chr3_18137155_18137306 | 0.65 |
Gm23686 |
predicted gene, 23686 |
40395 |
0.15 |
| chr8_82402195_82403147 | 0.59 |
Il15 |
interleukin 15 |
101 |
0.98 |
| chr7_126673507_126673658 | 0.58 |
Sgf29 |
SAGA complex associated factor 29 |
1551 |
0.16 |
| chr11_16761103_16761540 | 0.58 |
Egfr |
epidermal growth factor receptor |
9091 |
0.2 |
| chr6_149225033_149225369 | 0.58 |
1700003I16Rik |
RIKEN cDNA 1700003I16 gene |
10016 |
0.16 |
| chr9_74872523_74872674 | 0.56 |
Onecut1 |
one cut domain, family member 1 |
6114 |
0.16 |
| chr3_148786931_148787086 | 0.56 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
35971 |
0.22 |
| chr2_126934794_126934945 | 0.54 |
Sppl2a |
signal peptide peptidase like 2A |
1634 |
0.35 |
| chr2_148017296_148017455 | 0.54 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
20895 |
0.16 |
| chr17_89449780_89449962 | 0.54 |
Gm4719 |
predicted gene 4719 |
70630 |
0.13 |
| chr6_28491147_28491308 | 0.53 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
10798 |
0.14 |
| chr10_95254224_95254433 | 0.53 |
Gm48880 |
predicted gene, 48880 |
60525 |
0.08 |
| chr12_104083020_104083260 | 0.52 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr7_140719940_140720091 | 0.52 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
1267 |
0.28 |
| chr6_52606460_52606970 | 0.52 |
Gm44434 |
predicted gene, 44434 |
3470 |
0.19 |
| chr6_37748273_37748424 | 0.51 |
Atp6v0c-ps2 |
ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2 |
57738 |
0.12 |
| chr2_73491256_73491419 | 0.50 |
Wipf1 |
WAS/WASL interacting protein family, member 1 |
4868 |
0.19 |
| chr5_147312834_147312985 | 0.50 |
Cdx2 |
caudal type homeobox 2 |
5639 |
0.11 |
| chr19_40156899_40157166 | 0.49 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
30254 |
0.13 |
| chr2_73628966_73629117 | 0.49 |
Chn1 |
chimerin 1 |
3301 |
0.21 |
| chr3_116915063_116915225 | 0.47 |
Frrs1 |
ferric-chelate reductase 1 |
18434 |
0.13 |
| chr4_62016868_62017437 | 0.46 |
Mup-ps20 |
major urinary protein, pseudogene 20 |
2295 |
0.23 |
| chr5_148633016_148633167 | 0.45 |
Gm29815 |
predicted gene, 29815 |
19447 |
0.18 |
| chr5_125506718_125506869 | 0.45 |
Aacs |
acetoacetyl-CoA synthetase |
644 |
0.65 |
| chr1_107942218_107942404 | 0.45 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
5602 |
0.2 |
| chr10_121457902_121458053 | 0.45 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
4236 |
0.14 |
| chr3_116520495_116520697 | 0.44 |
Dbt |
dihydrolipoamide branched chain transacylase E2 |
268 |
0.83 |
| chr5_8667945_8668138 | 0.43 |
Gm42684 |
predicted gene 42684 |
1479 |
0.38 |
| chr7_120187015_120187182 | 0.43 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
13240 |
0.13 |
| chr3_152247820_152247987 | 0.42 |
Nexn |
nexilin |
361 |
0.8 |
| chr4_53222171_53222454 | 0.42 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
4455 |
0.21 |
| chr8_64837268_64837552 | 0.41 |
Gm45345 |
predicted gene 45345 |
824 |
0.55 |
| chr8_93190868_93191185 | 0.41 |
Gm45909 |
predicted gene 45909 |
332 |
0.84 |
| chr5_87562697_87562848 | 0.40 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
2581 |
0.17 |
| chr19_12654451_12654602 | 0.40 |
Gm24521 |
predicted gene, 24521 |
10729 |
0.09 |
| chr6_24049175_24049330 | 0.40 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
59319 |
0.12 |
| chr18_76968080_76968231 | 0.40 |
Hdhd2 |
haloacid dehalogenase-like hydrolase domain containing 2 |
23718 |
0.15 |
| chr17_15077386_15077778 | 0.39 |
Ermard |
ER membrane associated RNA degradation |
11967 |
0.15 |
| chr19_40141682_40141833 | 0.39 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
45529 |
0.11 |
| chr6_121171179_121171645 | 0.39 |
Pex26 |
peroxisomal biogenesis factor 26 |
12255 |
0.13 |
| chr2_122149958_122150187 | 0.39 |
B2m |
beta-2 microglobulin |
2386 |
0.19 |
| chr8_61543473_61543757 | 0.39 |
Palld |
palladin, cytoskeletal associated protein |
9774 |
0.25 |
| chr9_122847374_122847863 | 0.38 |
Gm47140 |
predicted gene, 47140 |
800 |
0.46 |
| chr15_67149186_67149444 | 0.38 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
20292 |
0.24 |
| chr6_94083257_94083610 | 0.38 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
169743 |
0.03 |
| chr7_134753489_134753670 | 0.38 |
Dock1 |
dedicator of cytokinesis 1 |
23876 |
0.24 |
| chr5_130007895_130008067 | 0.37 |
Gusb |
glucuronidase, beta |
4932 |
0.13 |
| chr6_149134216_149134396 | 0.37 |
Gm10203 |
predicted gene 10203 |
4136 |
0.14 |
| chr17_32944138_32944328 | 0.37 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
3102 |
0.14 |
| chr15_10192008_10192172 | 0.37 |
Prlr |
prolactin receptor |
14309 |
0.28 |
| chr2_180678351_180678502 | 0.37 |
Dido1 |
death inducer-obliterator 1 |
2471 |
0.19 |
| chr11_16821774_16821942 | 0.36 |
Egfros |
epidermal growth factor receptor, opposite strand |
8844 |
0.22 |
| chr2_180695831_180695982 | 0.36 |
Gm22502 |
predicted gene, 22502 |
2157 |
0.2 |
| chr11_17651863_17652036 | 0.35 |
Gm12016 |
predicted gene 12016 |
12766 |
0.3 |
| chr6_149150480_149150842 | 0.35 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
9015 |
0.13 |
| chr5_86920931_86921145 | 0.35 |
Gm25211 |
predicted gene, 25211 |
4716 |
0.11 |
| chr14_88428338_88428489 | 0.35 |
Gm48930 |
predicted gene, 48930 |
12388 |
0.18 |
| chr8_26351792_26351988 | 0.34 |
Gm31784 |
predicted gene, 31784 |
39556 |
0.12 |
| chr5_151394352_151394569 | 0.34 |
1700028E10Rik |
RIKEN cDNA 1700028E10 gene |
5684 |
0.19 |
| chr1_85325110_85325261 | 0.34 |
Gm16025 |
predicted gene 16025 |
1544 |
0.27 |
| chr10_68113055_68113212 | 0.34 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
23493 |
0.21 |
| chr7_90142238_90142404 | 0.34 |
Gm45222 |
predicted gene 45222 |
3627 |
0.14 |
| chr9_74894770_74895613 | 0.34 |
Onecut1 |
one cut domain, family member 1 |
28707 |
0.13 |
| chr19_10099300_10099508 | 0.34 |
Fads2 |
fatty acid desaturase 2 |
2342 |
0.23 |
| chr11_16798476_16798627 | 0.34 |
Egfros |
epidermal growth factor receptor, opposite strand |
32151 |
0.16 |
| chr11_99056309_99056460 | 0.33 |
Igfbp4 |
insulin-like growth factor binding protein 4 |
9073 |
0.12 |
| chr10_87545070_87545221 | 0.33 |
Pah |
phenylalanine hydroxylase |
1528 |
0.44 |
| chr12_57538162_57538324 | 0.33 |
Foxa1 |
forkhead box A1 |
7878 |
0.15 |
| chr9_122848440_122848788 | 0.33 |
Gm47140 |
predicted gene, 47140 |
196 |
0.89 |
| chr16_93346268_93346483 | 0.33 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
6814 |
0.19 |
| chr5_62748532_62748735 | 0.33 |
Arap2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
17487 |
0.24 |
| chr2_147998244_147998434 | 0.33 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
22154 |
0.18 |
| chr1_162963349_162963500 | 0.33 |
Gm37273 |
predicted gene, 37273 |
19590 |
0.14 |
| chr16_23326741_23326947 | 0.33 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
36374 |
0.13 |
| chr13_49644072_49644240 | 0.33 |
Cenpp |
centromere protein P |
8611 |
0.12 |
| chr3_57421025_57421339 | 0.32 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
4132 |
0.26 |
| chr10_95424636_95424797 | 0.32 |
5730420D15Rik |
RIKEN cDNA 5730420D15 gene |
7341 |
0.13 |
| chr10_68121308_68121503 | 0.32 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
15221 |
0.24 |
| chr11_16829553_16829792 | 0.32 |
Egfros |
epidermal growth factor receptor, opposite strand |
1030 |
0.59 |
| chr11_72631760_72631911 | 0.32 |
Gm24143 |
predicted gene, 24143 |
15685 |
0.13 |
| chr5_90457553_90457769 | 0.32 |
Alb |
albumin |
3236 |
0.21 |
| chr10_87923338_87923500 | 0.32 |
Tyms-ps |
thymidylate synthase, pseudogene |
43428 |
0.12 |
| chr14_34328663_34328894 | 0.32 |
Glud1 |
glutamate dehydrogenase 1 |
116 |
0.93 |
| chr2_73939236_73939432 | 0.31 |
Gm13668 |
predicted gene 13668 |
21335 |
0.17 |
| chr18_9487035_9487277 | 0.31 |
Gm7527 |
predicted gene 7527 |
8465 |
0.16 |
| chr1_24116155_24116306 | 0.31 |
Gm26607 |
predicted gene, 26607 |
9045 |
0.17 |
| chr18_33352007_33352191 | 0.31 |
Gm5503 |
predicted gene 5503 |
32856 |
0.21 |
| chr7_63899068_63899509 | 0.31 |
Gm27252 |
predicted gene 27252 |
1314 |
0.37 |
| chr17_28428613_28428954 | 0.31 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr17_64720655_64720806 | 0.30 |
Man2a1 |
mannosidase 2, alpha 1 |
7111 |
0.22 |
| chr17_46053621_46054045 | 0.30 |
Vegfa |
vascular endothelial growth factor A |
21464 |
0.12 |
| chr2_121439848_121440228 | 0.30 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
1113 |
0.25 |
| chr1_127898839_127898990 | 0.30 |
Rab3gap1 |
RAB3 GTPase activating protein subunit 1 |
1999 |
0.31 |
| chr5_36621044_36621360 | 0.30 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
53 |
0.96 |
| chrX_139665962_139666117 | 0.30 |
Gm6322 |
predicted gene 6322 |
11450 |
0.17 |
| chr1_126431765_126431942 | 0.30 |
Nckap5 |
NCK-associated protein 5 |
16603 |
0.29 |
| chr14_35502504_35502655 | 0.30 |
Gm19016 |
predicted gene, 19016 |
46547 |
0.17 |
| chr7_72323573_72323742 | 0.30 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
17049 |
0.28 |
| chr16_86389664_86389844 | 0.30 |
Gm32357 |
predicted gene, 32357 |
153583 |
0.04 |
| chr4_57122143_57122466 | 0.30 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
2244 |
0.35 |
| chr15_4678897_4679270 | 0.30 |
C6 |
complement component 6 |
48092 |
0.17 |
| chr9_111128128_111128279 | 0.29 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
8528 |
0.16 |
| chr14_122876950_122877397 | 0.29 |
Pcca |
propionyl-Coenzyme A carboxylase, alpha polypeptide |
1238 |
0.47 |
| chr2_109688795_109688948 | 0.29 |
Bdnf |
brain derived neurotrophic factor |
3565 |
0.23 |
| chr13_85208789_85208956 | 0.29 |
Ccnh |
cyclin H |
2450 |
0.3 |
| chr5_125528759_125528910 | 0.29 |
Tmem132b |
transmembrane protein 132B |
2940 |
0.22 |
| chr11_104387417_104387568 | 0.29 |
Gm47315 |
predicted gene, 47315 |
37808 |
0.13 |
| chr8_105009723_105009901 | 0.29 |
Ces2h |
carboxylesterase 2H |
8959 |
0.1 |
| chr8_90871765_90872190 | 0.29 |
Gm45640 |
predicted gene 45640 |
4604 |
0.15 |
| chr9_35123533_35123708 | 0.29 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
4352 |
0.15 |
| chr14_13625050_13625231 | 0.29 |
Sntn |
sentan, cilia apical structure protein |
45736 |
0.17 |
| chr3_79555181_79555357 | 0.29 |
Gm3513 |
predicted gene 3513 |
3076 |
0.17 |
| chr6_149359539_149359690 | 0.29 |
Gm15784 |
predicted gene 15784 |
2072 |
0.23 |
| chr11_29526849_29527005 | 0.29 |
Mtif2 |
mitochondrial translational initiation factor 2 |
470 |
0.69 |
| chr11_7815566_7815774 | 0.29 |
Gm27393 |
predicted gene, 27393 |
70835 |
0.13 |
| chr18_64079171_64079335 | 0.29 |
Gm6974 |
predicted gene 6974 |
2167 |
0.36 |
| chr2_42039001_42039161 | 0.28 |
Gm13461 |
predicted gene 13461 |
50252 |
0.18 |
| chr5_67300787_67300957 | 0.28 |
Slc30a9 |
solute carrier family 30 (zinc transporter), member 9 |
6083 |
0.19 |
| chr6_108466356_108466533 | 0.28 |
Gm44101 |
predicted gene, 44101 |
6131 |
0.18 |
| chr17_32935826_32936018 | 0.28 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
4914 |
0.11 |
| chr12_70196878_70197068 | 0.28 |
Pygl |
liver glycogen phosphorylase |
868 |
0.54 |
| chr11_60201956_60202762 | 0.28 |
Mir6922 |
microRNA 6922 |
32 |
0.63 |
| chr18_12663657_12663965 | 0.28 |
Gm41668 |
predicted gene, 41668 |
15392 |
0.14 |
| chr15_3476580_3476767 | 0.28 |
Ghr |
growth hormone receptor |
5029 |
0.32 |
| chr7_44815923_44816372 | 0.28 |
Nup62 |
nucleoporin 62 |
59 |
0.66 |
| chr13_23804366_23804536 | 0.28 |
Gm11337 |
predicted gene 11337 |
148 |
0.87 |
| chr2_32095451_32095626 | 0.27 |
Plpp7 |
phospholipid phosphatase 7 (inactive) |
10 |
0.96 |
| chr6_14778417_14778568 | 0.27 |
Ppp1r3a |
protein phosphatase 1, regulatory subunit 3A |
23218 |
0.27 |
| chr8_82524759_82525044 | 0.27 |
Gm8167 |
predicted gene 8167 |
52866 |
0.15 |
| chr4_8688195_8688404 | 0.27 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
2107 |
0.38 |
| chr1_23279889_23280199 | 0.27 |
Mir30a |
microRNA 30a |
7775 |
0.12 |
| chr11_16807603_16807776 | 0.27 |
Egfros |
epidermal growth factor receptor, opposite strand |
23013 |
0.19 |
| chr1_44575664_44575838 | 0.27 |
Gm37626 |
predicted gene, 37626 |
14866 |
0.19 |
| chr3_14685860_14686011 | 0.27 |
Gm5843 |
predicted gene 5843 |
36481 |
0.12 |
| chr10_123842451_123842602 | 0.27 |
Gm18510 |
predicted gene, 18510 |
49372 |
0.19 |
| chr3_28591458_28591833 | 0.27 |
Tnik |
TRAF2 and NCK interacting kinase |
38227 |
0.18 |
| chr1_107716693_107716857 | 0.27 |
Gm15389 |
predicted gene 15389 |
29767 |
0.18 |
| chr2_4998266_4998605 | 0.27 |
Mcm10 |
minichromosome maintenance 10 replication initiation factor |
2541 |
0.2 |
| chr1_33939932_33940113 | 0.27 |
Gm37354 |
predicted gene, 37354 |
17567 |
0.12 |
| chr11_71013317_71013604 | 0.27 |
Derl2 |
Der1-like domain family, member 2 |
5752 |
0.1 |
| chr1_170137562_170137720 | 0.26 |
Uap1 |
UDP-N-acetylglucosamine pyrophosphorylase 1 |
6072 |
0.16 |
| chr1_92944221_92944381 | 0.26 |
Capn10 |
calpain 10 |
1597 |
0.22 |
| chr4_99037726_99038187 | 0.26 |
Angptl3 |
angiopoietin-like 3 |
4753 |
0.21 |
| chr15_75205125_75205276 | 0.26 |
Gm28117 |
predicted gene 28117 |
7533 |
0.12 |
| chr13_24144521_24144672 | 0.26 |
Carmil1 |
capping protein regulator and myosin 1 linker 1 |
10613 |
0.19 |
| chr11_11845100_11845416 | 0.26 |
Ddc |
dopa decarboxylase |
8978 |
0.18 |
| chr6_28734300_28734519 | 0.26 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
28689 |
0.19 |
| chr15_58971828_58971979 | 0.26 |
Mtss1 |
MTSS I-BAR domain containing 1 |
636 |
0.67 |
| chr8_93173439_93173590 | 0.26 |
Ces1d |
carboxylesterase 1D |
1775 |
0.27 |
| chr5_130020282_130020619 | 0.26 |
Asl |
argininosuccinate lyase |
3850 |
0.14 |
| chr2_170269712_170270084 | 0.26 |
Gm14270 |
predicted gene 14270 |
15137 |
0.23 |
| chr4_76360115_76360401 | 0.26 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
16015 |
0.23 |
| chr19_36323375_36323536 | 0.25 |
Gm47735 |
predicted gene, 47735 |
24843 |
0.13 |
| chr11_78846353_78846604 | 0.25 |
Lyrm9 |
LYR motif containing 9 |
19849 |
0.15 |
| chr5_147224671_147224829 | 0.25 |
Gm24556 |
predicted gene, 24556 |
24497 |
0.11 |
| chr4_150709744_150709942 | 0.25 |
Gm16079 |
predicted gene 16079 |
31051 |
0.16 |
| chr17_14962228_14962437 | 0.25 |
Phf10 |
PHD finger protein 10 |
1059 |
0.36 |
| chr7_73557395_73557719 | 0.25 |
1810026B05Rik |
RIKEN cDNA 1810026B05 gene |
416 |
0.74 |
| chr2_60288667_60288955 | 0.25 |
Cd302 |
CD302 antigen |
4323 |
0.19 |
| chr11_111297916_111298094 | 0.25 |
Gm11675 |
predicted gene 11675 |
21905 |
0.28 |
| chr4_19491208_19491365 | 0.25 |
Cpne3 |
copine III |
52392 |
0.15 |
| chr1_136140658_136140838 | 0.25 |
Kif21b |
kinesin family member 21B |
9294 |
0.11 |
| chr3_18877514_18877665 | 0.25 |
Gm30341 |
predicted gene, 30341 |
124512 |
0.06 |
| chr6_83922439_83922590 | 0.25 |
Zfp638 |
zinc finger protein 638 |
6341 |
0.13 |
| chr8_9027774_9027937 | 0.25 |
Gm44515 |
predicted gene 44515 |
95156 |
0.06 |
| chr4_44784029_44784206 | 0.25 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
23673 |
0.15 |
| chr6_71423223_71423388 | 0.24 |
Gm44130 |
predicted gene, 44130 |
11802 |
0.1 |
| chr2_134500653_134500804 | 0.24 |
Hao1 |
hydroxyacid oxidase 1, liver |
53579 |
0.17 |
| chr11_82032806_82032957 | 0.24 |
Ccl2 |
chemokine (C-C motif) ligand 2 |
2690 |
0.17 |
| chr1_159231811_159232536 | 0.24 |
Cop1 |
COP1, E3 ubiquitin ligase |
147 |
0.95 |
| chr6_89381990_89382152 | 0.24 |
Plxna1 |
plexin A1 |
19451 |
0.17 |
| chr14_58710319_58710470 | 0.24 |
Gm25614 |
predicted gene, 25614 |
59994 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.2 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.6 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.1 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.3 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0048369 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.4 | GO:0052696 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.6 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.0 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.0 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.0 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.0 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.5 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.0 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0018734 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.0 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0034834 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |