Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
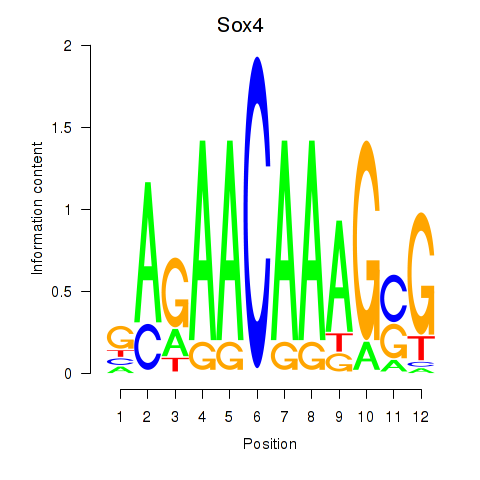
Results for Sox4
Z-value: 1.00
Transcription factors associated with Sox4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox4
|
ENSMUSG00000076431.4 | SRY (sex determining region Y)-box 4 |
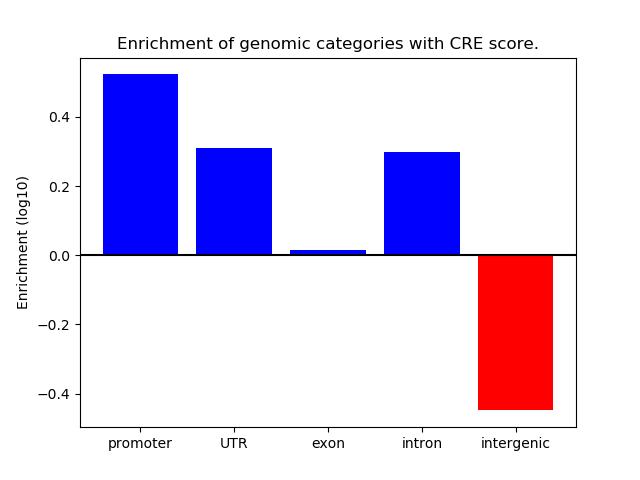
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_28930434_28930607 | Sox4 | 23193 | 0.167201 | 0.82 | 4.5e-02 | Click! |
| chr13_28958185_28958642 | Sox4 | 4700 | 0.246141 | -0.80 | 5.8e-02 | Click! |
| chr13_28958754_28958905 | Sox4 | 5116 | 0.241122 | -0.51 | 3.1e-01 | Click! |
| chr13_28953339_28953518 | Sox4 | 285 | 0.922547 | -0.49 | 3.2e-01 | Click! |
| chr13_28957891_28958075 | Sox4 | 4270 | 0.252776 | -0.41 | 4.2e-01 | Click! |
Activity of the Sox4 motif across conditions
Conditions sorted by the z-value of the Sox4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
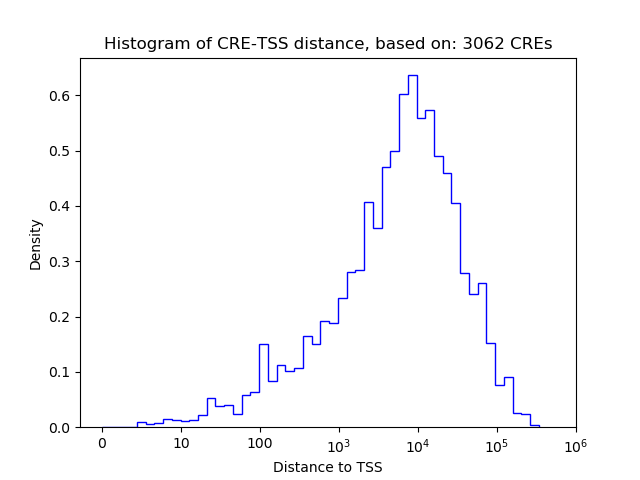
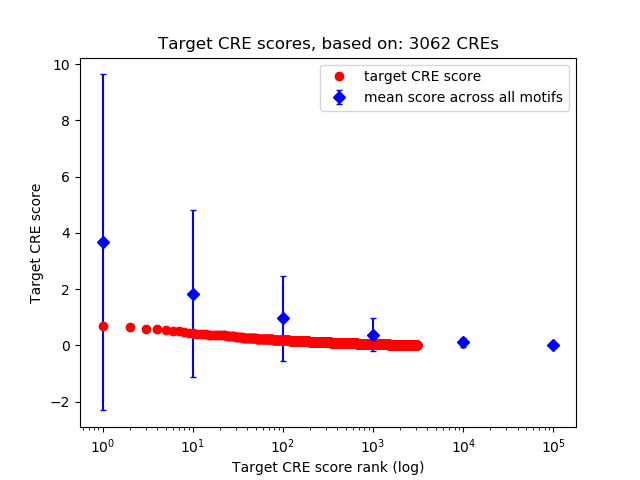
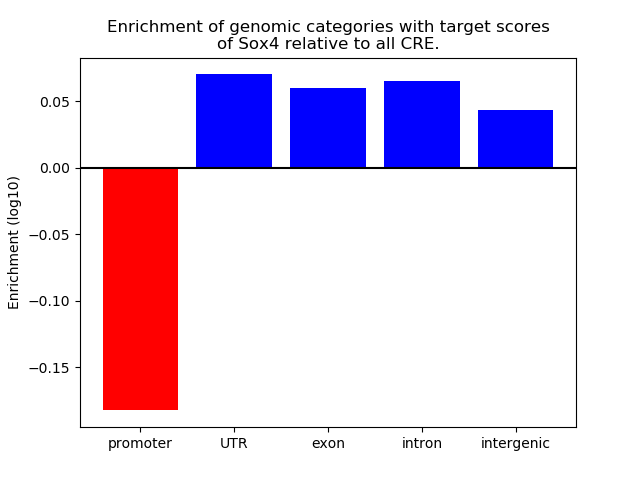
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_83244234_83244393 | 0.70 |
Slc41a2 |
solute carrier family 41, member 2 |
60062 |
0.11 |
| chr7_113935146_113935310 | 0.64 |
Gm45615 |
predicted gene 45615 |
151670 |
0.04 |
| chr4_35916085_35916245 | 0.59 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
70961 |
0.13 |
| chr7_24500290_24500448 | 0.58 |
Cadm4 |
cell adhesion molecule 4 |
408 |
0.68 |
| chr19_16182027_16182207 | 0.53 |
E030024N20Rik |
RIKEN cDNA E030024N20 gene |
17312 |
0.19 |
| chr7_66106744_66106923 | 0.52 |
Chsy1 |
chondroitin sulfate synthase 1 |
2682 |
0.17 |
| chr2_58771297_58771448 | 0.52 |
Upp2 |
uridine phosphorylase 2 |
6047 |
0.22 |
| chr8_13169044_13169201 | 0.46 |
Lamp1 |
lysosomal-associated membrane protein 1 |
4059 |
0.12 |
| chr15_74994754_74994914 | 0.45 |
Ly6a |
lymphocyte antigen 6 complex, locus A |
2800 |
0.12 |
| chr5_118571610_118571780 | 0.44 |
Gm43785 |
predicted gene 43785 |
8594 |
0.15 |
| chr4_130749359_130749859 | 0.41 |
Snord85 |
small nucleolar RNA, C/D box 85 |
25 |
0.96 |
| chr7_132585908_132586066 | 0.40 |
Oat |
ornithine aminotransferase |
9589 |
0.14 |
| chr9_74892937_74893252 | 0.39 |
Onecut1 |
one cut domain, family member 1 |
26610 |
0.14 |
| chr7_26829282_26829437 | 0.39 |
Cyp2a5 |
cytochrome P450, family 2, subfamily a, polypeptide 5 |
5946 |
0.17 |
| chr2_32484947_32485331 | 0.38 |
Gm37169 |
predicted gene, 37169 |
2849 |
0.15 |
| chr14_73161911_73162182 | 0.38 |
Rcbtb2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
106 |
0.96 |
| chr3_98047351_98047638 | 0.38 |
Gm42819 |
predicted gene 42819 |
16807 |
0.17 |
| chr18_38962486_38962657 | 0.37 |
Gm5820 |
predicted gene 5820 |
6862 |
0.19 |
| chr2_15610043_15610194 | 0.37 |
Gm37595 |
predicted gene, 37595 |
79005 |
0.1 |
| chr19_10067201_10067465 | 0.35 |
Fads2 |
fatty acid desaturase 2 |
3854 |
0.16 |
| chr11_16822583_16822870 | 0.35 |
Egfros |
epidermal growth factor receptor, opposite strand |
7976 |
0.23 |
| chr2_17342233_17342437 | 0.35 |
Nebl |
nebulette |
18751 |
0.27 |
| chr18_84376797_84376986 | 0.35 |
Gm37216 |
predicted gene, 37216 |
745 |
0.73 |
| chr5_125521579_125521730 | 0.35 |
Aacs |
acetoacetyl-CoA synthetase |
6411 |
0.16 |
| chr18_65520606_65520833 | 0.35 |
Gm30018 |
predicted gene, 30018 |
11082 |
0.12 |
| chr16_97792461_97792626 | 0.34 |
Prdm15 |
PR domain containing 15 |
5376 |
0.19 |
| chr6_108513973_108514142 | 0.34 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
4675 |
0.17 |
| chr18_81199291_81199673 | 0.34 |
4930594M17Rik |
RIKEN cDNA 4930594M17 gene |
36584 |
0.17 |
| chr7_80335806_80335957 | 0.33 |
Unc45a |
unc-45 myosin chaperone A |
3685 |
0.12 |
| chr2_75960594_75960799 | 0.30 |
Ttc30a2 |
tetratricopeptide repeat domain 30A2 |
17474 |
0.14 |
| chr11_98767825_98767976 | 0.29 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2526 |
0.15 |
| chr4_54733562_54733749 | 0.29 |
Gm12477 |
predicted gene 12477 |
63463 |
0.11 |
| chr9_96437861_96438072 | 0.28 |
BC043934 |
cDNA sequence BC043934 |
172 |
0.94 |
| chr6_141662549_141662700 | 0.28 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
10369 |
0.27 |
| chr7_19441871_19442030 | 0.28 |
Mark4 |
MAP/microtubule affinity regulating kinase 4 |
6666 |
0.08 |
| chr3_148957664_148957815 | 0.27 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
2952 |
0.29 |
| chr7_15981709_15981860 | 0.27 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
3059 |
0.15 |
| chr19_34828291_34828506 | 0.27 |
Mir107 |
microRNA 107 |
7625 |
0.14 |
| chr11_98775849_98776027 | 0.26 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
605 |
0.56 |
| chr5_147313676_147313887 | 0.26 |
Cdx2 |
caudal type homeobox 2 |
6511 |
0.11 |
| chr12_54710441_54710776 | 0.26 |
Gm22634 |
predicted gene, 22634 |
300 |
0.72 |
| chr11_16818941_16819203 | 0.26 |
Egfros |
epidermal growth factor receptor, opposite strand |
11630 |
0.22 |
| chrX_159817258_159817435 | 0.26 |
Sh3kbp1 |
SH3-domain kinase binding protein 1 |
23122 |
0.25 |
| chr5_130027278_130027450 | 0.25 |
Asl |
argininosuccinate lyase |
1883 |
0.2 |
| chr4_130630552_130630730 | 0.25 |
Pum1 |
pumilio RNA-binding family member 1 |
32680 |
0.16 |
| chr17_63010946_63011130 | 0.25 |
Gm25348 |
predicted gene, 25348 |
85962 |
0.1 |
| chr7_117386572_117386727 | 0.25 |
Gm45153 |
predicted gene 45153 |
4787 |
0.26 |
| chr9_13681735_13681905 | 0.25 |
Maml2 |
mastermind like transcriptional coactivator 2 |
19360 |
0.18 |
| chr19_56170856_56171017 | 0.25 |
Gm31912 |
predicted gene, 31912 |
64626 |
0.11 |
| chr16_20412097_20412250 | 0.24 |
Abcc5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
11887 |
0.14 |
| chr2_65441659_65441885 | 0.24 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
53459 |
0.11 |
| chr8_104095732_104095883 | 0.24 |
Cdh5 |
cadherin 5 |
5818 |
0.15 |
| chr1_85093829_85093980 | 0.23 |
Gm10553 |
predicted gene 10553 |
5608 |
0.1 |
| chr11_16854764_16854960 | 0.23 |
Egfr |
epidermal growth factor receptor |
23288 |
0.17 |
| chr19_10094218_10094369 | 0.23 |
Fads2 |
fatty acid desaturase 2 |
5577 |
0.16 |
| chr11_16813698_16813849 | 0.23 |
Egfros |
epidermal growth factor receptor, opposite strand |
16929 |
0.21 |
| chr2_141285965_141286164 | 0.23 |
Macrod2os1 |
mono-ADP ribosylhydrolase 2, opposite strand 1 |
927 |
0.69 |
| chr1_164120420_164120571 | 0.23 |
Selp |
selectin, platelet |
5231 |
0.14 |
| chr7_50105525_50105881 | 0.23 |
Nell1 |
NEL-like 1 |
7393 |
0.32 |
| chr1_178528531_178528699 | 0.23 |
Kif26b |
kinesin family member 26B |
510 |
0.84 |
| chr7_79309461_79309624 | 0.23 |
Gm39041 |
predicted gene, 39041 |
10517 |
0.14 |
| chr1_36807833_36808005 | 0.22 |
Tmem131 |
transmembrane protein 131 |
2617 |
0.21 |
| chr2_118696590_118696966 | 0.22 |
Ankrd63 |
ankyrin repeat domain 63 |
7185 |
0.13 |
| chr7_26301156_26301307 | 0.22 |
Cyp2a4 |
cytochrome P450, family 2, subfamily a, polypeptide 4 |
5938 |
0.13 |
| chr17_10511158_10511318 | 0.22 |
Gm16168 |
predicted gene 16168 |
14785 |
0.24 |
| chr16_10976433_10976649 | 0.22 |
Litaf |
LPS-induced TN factor |
962 |
0.41 |
| chr1_165184250_165184426 | 0.21 |
Sft2d2 |
SFT2 domain containing 2 |
9979 |
0.14 |
| chr6_141671866_141672017 | 0.21 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
9305 |
0.27 |
| chr14_76805432_76805625 | 0.21 |
Gm30246 |
predicted gene, 30246 |
24701 |
0.16 |
| chr7_65467465_65467748 | 0.21 |
Gm44792 |
predicted gene 44792 |
13615 |
0.21 |
| chr2_134547439_134547602 | 0.21 |
Hao1 |
hydroxyacid oxidase 1, liver |
6787 |
0.31 |
| chr2_132595669_132595997 | 0.21 |
AU019990 |
expressed sequence AU019990 |
2362 |
0.24 |
| chr11_16857621_16857825 | 0.21 |
Egfr |
epidermal growth factor receptor |
20427 |
0.18 |
| chr1_59481889_59482258 | 0.21 |
Fzd7 |
frizzled class receptor 7 |
351 |
0.83 |
| chr2_38531241_38531405 | 0.21 |
Gm35808 |
predicted gene, 35808 |
4384 |
0.14 |
| chr7_99848918_99849257 | 0.21 |
Spcs2 |
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
9772 |
0.11 |
| chr2_68874298_68874495 | 0.21 |
Cers6 |
ceramide synthase 6 |
12810 |
0.14 |
| chr18_75389536_75389700 | 0.20 |
Smad7 |
SMAD family member 7 |
14704 |
0.21 |
| chr17_69982645_69982841 | 0.20 |
Dlgap1 |
DLG associated protein 1 |
13322 |
0.24 |
| chr9_98429903_98430078 | 0.20 |
Rbp1 |
retinol binding protein 1, cellular |
7029 |
0.21 |
| chr17_47304154_47304305 | 0.20 |
Trerf1 |
transcriptional regulating factor 1 |
10160 |
0.16 |
| chr5_135725445_135725806 | 0.20 |
Por |
P450 (cytochrome) oxidoreductase |
103 |
0.94 |
| chr7_98002722_98002888 | 0.20 |
Gdpd4 |
glycerophosphodiester phosphodiesterase domain containing 4 |
44984 |
0.13 |
| chr10_94469219_94469370 | 0.20 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
45563 |
0.14 |
| chr2_42022841_42022992 | 0.20 |
Gm13461 |
predicted gene 13461 |
34087 |
0.23 |
| chr19_12718913_12719287 | 0.20 |
Gm15962 |
predicted gene 15962 |
2103 |
0.19 |
| chr19_8613413_8613628 | 0.19 |
Slc22a6 |
solute carrier family 22 (organic anion transporter), member 6 |
4519 |
0.12 |
| chr1_193928644_193928965 | 0.19 |
Gm21362 |
predicted gene, 21362 |
61795 |
0.15 |
| chr4_97765098_97765412 | 0.19 |
Nfia |
nuclear factor I/A |
7479 |
0.2 |
| chr8_11010925_11011239 | 0.19 |
Irs2 |
insulin receptor substrate 2 |
2624 |
0.19 |
| chr4_154506853_154507016 | 0.19 |
Prdm16 |
PR domain containing 16 |
21831 |
0.18 |
| chr5_54034513_54034730 | 0.19 |
Stim2 |
stromal interaction molecule 2 |
36056 |
0.18 |
| chr9_15537781_15537949 | 0.19 |
Smco4 |
single-pass membrane protein with coiled-coil domains 4 |
17006 |
0.16 |
| chr9_74706331_74706505 | 0.19 |
Gm27233 |
predicted gene 27233 |
2844 |
0.32 |
| chr1_170924605_170924775 | 0.19 |
Gm2962 |
predicted pseudogene 2962 |
954 |
0.33 |
| chr10_80174407_80174582 | 0.19 |
Fam174c |
family with sequence similarity 174, member C |
1550 |
0.18 |
| chr6_66898216_66898367 | 0.19 |
Gng12 |
guanine nucleotide binding protein (G protein), gamma 12 |
1400 |
0.3 |
| chr11_16872747_16873031 | 0.18 |
Egfr |
epidermal growth factor receptor |
5261 |
0.23 |
| chr14_22589711_22589891 | 0.18 |
Lrmda |
leucine rich melanocyte differentiation associated |
6712 |
0.24 |
| chr1_165817347_165817510 | 0.18 |
Gm23402 |
predicted gene, 23402 |
7686 |
0.1 |
| chr4_49553429_49553778 | 0.18 |
Aldob |
aldolase B, fructose-bisphosphate |
4057 |
0.17 |
| chr2_158037741_158037902 | 0.18 |
Rprd1b |
regulation of nuclear pre-mRNA domain containing 1B |
5602 |
0.18 |
| chr11_16784544_16784843 | 0.18 |
Egfr |
epidermal growth factor receptor |
32463 |
0.16 |
| chr3_79903884_79904035 | 0.18 |
Gm36569 |
predicted gene, 36569 |
17153 |
0.17 |
| chr9_6786497_6786659 | 0.18 |
Gm48675 |
predicted gene, 48675 |
15265 |
0.26 |
| chr11_70236702_70236853 | 0.18 |
0610010K14Rik |
RIKEN cDNA 0610010K14 gene |
127 |
0.86 |
| chr3_89139844_89139995 | 0.18 |
Pklr |
pyruvate kinase liver and red blood cell |
3296 |
0.09 |
| chr3_9610870_9611024 | 0.18 |
Zfp704 |
zinc finger protein 704 |
862 |
0.67 |
| chr2_129526434_129526601 | 0.18 |
F830045P16Rik |
RIKEN cDNA F830045P16 gene |
10085 |
0.16 |
| chr8_46387074_46387264 | 0.18 |
Gm45253 |
predicted gene 45253 |
1281 |
0.38 |
| chr6_55170633_55170787 | 0.18 |
Inmt |
indolethylamine N-methyltransferase |
4304 |
0.19 |
| chr14_66132883_66133034 | 0.18 |
Chrna2 |
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
2081 |
0.27 |
| chr14_119816401_119816559 | 0.18 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
17502 |
0.21 |
| chr6_149344317_149344633 | 0.18 |
Gm15782 |
predicted gene 15782 |
9351 |
0.14 |
| chr16_96127404_96127899 | 0.17 |
Hmgn1 |
high mobility group nucleosomal binding domain 1 |
47 |
0.96 |
| chr8_72503838_72503991 | 0.17 |
Gm17435 |
predicted gene, 17435 |
10970 |
0.08 |
| chr17_6018338_6018665 | 0.17 |
Synj2 |
synaptojanin 2 |
4952 |
0.19 |
| chr15_85538329_85538480 | 0.17 |
Gm4825 |
predicted pseudogene 4825 |
27592 |
0.13 |
| chr13_73320522_73320699 | 0.17 |
Gm10263 |
predicted gene 10263 |
2766 |
0.2 |
| chr11_90356770_90356930 | 0.17 |
Hlf |
hepatic leukemia factor |
25085 |
0.22 |
| chr3_149239531_149239709 | 0.17 |
Gm10287 |
predicted gene 10287 |
13875 |
0.2 |
| chr8_107206262_107206573 | 0.16 |
Cyb5b |
cytochrome b5 type B |
40502 |
0.12 |
| chr1_134173185_134173337 | 0.16 |
Chil1 |
chitinase-like 1 |
8915 |
0.14 |
| chr8_107117129_107117285 | 0.16 |
C630050I24Rik |
RIKEN cDNA C630050I24 gene |
1903 |
0.22 |
| chr16_17656326_17656735 | 0.16 |
Med15 |
mediator complex subunit 15 |
677 |
0.52 |
| chr12_119071015_119071241 | 0.16 |
Gm18921 |
predicted gene, 18921 |
3698 |
0.34 |
| chr7_79279429_79279594 | 0.16 |
Gm44639 |
predicted gene 44639 |
1744 |
0.27 |
| chr17_87377697_87377853 | 0.16 |
Ttc7 |
tetratricopeptide repeat domain 7 |
1078 |
0.43 |
| chr14_60993572_60993791 | 0.16 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
17796 |
0.22 |
| chr12_110499327_110499478 | 0.16 |
Gm19605 |
predicted gene, 19605 |
13194 |
0.16 |
| chr9_72260390_72260691 | 0.16 |
Gm25163 |
predicted gene, 25163 |
3401 |
0.12 |
| chr9_121435320_121435493 | 0.16 |
Trak1 |
trafficking protein, kinesin binding 1 |
18890 |
0.17 |
| chr10_78452749_78452929 | 0.16 |
Pdxk |
pyridoxal (pyridoxine, vitamin B6) kinase |
4593 |
0.08 |
| chr3_18148711_18148862 | 0.16 |
Gm23686 |
predicted gene, 23686 |
28839 |
0.19 |
| chr2_177925591_177925999 | 0.16 |
Zfp972 |
zinc finger protein 972 |
182 |
0.93 |
| chr7_69979793_69979981 | 0.16 |
Gm24120 |
predicted gene, 24120 |
21983 |
0.21 |
| chr2_117129250_117129573 | 0.16 |
Spred1 |
sprouty protein with EVH-1 domain 1, related sequence |
7773 |
0.22 |
| chr4_108043471_108043786 | 0.16 |
Gm23354 |
predicted gene, 23354 |
1828 |
0.25 |
| chr7_112788334_112788697 | 0.16 |
Tead1 |
TEA domain family member 1 |
28969 |
0.19 |
| chr19_29470771_29470965 | 0.16 |
Pdcd1lg2 |
programmed cell death 1 ligand 2 |
48809 |
0.1 |
| chr19_21829238_21829531 | 0.16 |
Gm50130 |
predicted gene, 50130 |
4372 |
0.26 |
| chr15_25677510_25677678 | 0.16 |
Myo10 |
myosin X |
3084 |
0.24 |
| chr14_19986275_19986426 | 0.16 |
Gng2 |
guanine nucleotide binding protein (G protein), gamma 2 |
8723 |
0.21 |
| chr17_12658996_12659172 | 0.15 |
Slc22a1 |
solute carrier family 22 (organic cation transporter), member 1 |
6376 |
0.19 |
| chr1_39813213_39813385 | 0.15 |
Gm3646 |
predicted gene 3646 |
7967 |
0.21 |
| chr11_117234314_117234465 | 0.15 |
Septin9 |
septin 9 |
2104 |
0.3 |
| chr3_101804488_101804639 | 0.15 |
Mab21l3 |
mab-21-like 3 |
31660 |
0.17 |
| chr17_79020514_79020830 | 0.15 |
Prkd3 |
protein kinase D3 |
105 |
0.96 |
| chr7_26880928_26881079 | 0.15 |
Cyp2a21-ps |
cytochrome P450, family 2, subfamily a, polypeptide 21, pseudogene |
36378 |
0.11 |
| chr7_123405689_123405849 | 0.15 |
Lcmt1 |
leucine carboxyl methyltransferase 1 |
2411 |
0.28 |
| chr6_90436303_90436648 | 0.15 |
Cfap100 |
cilia and flagella associated protein 100 |
7678 |
0.12 |
| chr8_123702859_123703051 | 0.15 |
6030466F02Rik |
RIKEN cDNA 6030466F02 gene |
31003 |
0.05 |
| chr13_54632857_54633027 | 0.15 |
Faf2 |
Fas associated factor family member 2 |
8601 |
0.12 |
| chr19_40156899_40157166 | 0.15 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
30254 |
0.13 |
| chr16_32853567_32853731 | 0.15 |
Rubcn |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
14689 |
0.12 |
| chr14_60635934_60636102 | 0.15 |
Spata13 |
spermatogenesis associated 13 |
1263 |
0.52 |
| chr2_24864822_24864973 | 0.15 |
Ehmt1 |
euchromatic histone methyltransferase 1 |
974 |
0.44 |
| chrX_74245766_74245917 | 0.15 |
Flna |
filamin, alpha |
523 |
0.54 |
| chrX_111648183_111648334 | 0.15 |
Hdx |
highly divergent homeobox |
48821 |
0.16 |
| chr18_53744559_53744906 | 0.15 |
Cep120 |
centrosomal protein 120 |
124 |
0.98 |
| chr1_21267631_21267791 | 0.15 |
Gm28836 |
predicted gene 28836 |
3882 |
0.13 |
| chr6_38823949_38824290 | 0.15 |
Hipk2 |
homeodomain interacting protein kinase 2 |
5773 |
0.25 |
| chr4_57250984_57251161 | 0.15 |
Ptpn3 |
protein tyrosine phosphatase, non-receptor type 3 |
29018 |
0.17 |
| chr11_111602165_111602329 | 0.15 |
Gm11676 |
predicted gene 11676 |
11059 |
0.3 |
| chr11_121258796_121258982 | 0.15 |
Foxk2 |
forkhead box K2 |
1101 |
0.39 |
| chr17_66708120_66708311 | 0.15 |
1700016K05Rik |
RIKEN cDNA 1700016K05 gene |
6239 |
0.15 |
| chr17_79883308_79883515 | 0.15 |
Atl2 |
atlastin GTPase 2 |
12640 |
0.17 |
| chr11_45533343_45533513 | 0.15 |
Gm12162 |
predicted gene 12162 |
64924 |
0.12 |
| chrX_20547825_20547979 | 0.15 |
Rgn |
regucalcin |
1885 |
0.26 |
| chr13_102698753_102698921 | 0.14 |
Cd180 |
CD180 antigen |
5226 |
0.24 |
| chr4_139186254_139186405 | 0.14 |
Gm16287 |
predicted gene 16287 |
5674 |
0.15 |
| chr6_91614903_91615054 | 0.14 |
Gm45218 |
predicted gene 45218 |
15746 |
0.13 |
| chr7_141436148_141436439 | 0.14 |
Slc25a22 |
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
896 |
0.28 |
| chr9_9128366_9128693 | 0.14 |
Gm16833 |
predicted gene, 16833 |
107759 |
0.07 |
| chr3_138304273_138304430 | 0.14 |
Gm16559 |
predicted gene 16559 |
2671 |
0.17 |
| chr1_74968300_74968461 | 0.14 |
Gm37744 |
predicted gene, 37744 |
14128 |
0.12 |
| chr17_46016325_46016519 | 0.14 |
Vegfa |
vascular endothelial growth factor A |
4950 |
0.19 |
| chr17_29579809_29579984 | 0.14 |
Tbc1d22b |
TBC1 domain family, member 22B |
7473 |
0.09 |
| chr16_8829001_8829532 | 0.14 |
1810013L24Rik |
RIKEN cDNA 1810013L24 gene |
834 |
0.6 |
| chr2_91750560_91750711 | 0.14 |
Ambra1 |
autophagy/beclin 1 regulator 1 |
15810 |
0.14 |
| chr19_7788761_7788946 | 0.14 |
Slc22a26 |
solute carrier family 22 (organic cation transporter), member 26 |
13725 |
0.17 |
| chr6_86670951_86671106 | 0.14 |
Mxd1 |
MAX dimerization protein 1 |
1867 |
0.2 |
| chr7_113059372_113059769 | 0.14 |
Gm23662 |
predicted gene, 23662 |
60128 |
0.11 |
| chr11_4216611_4216764 | 0.14 |
Castor1 |
cytosolic arginine sensor for mTORC1 subunit 1 |
1538 |
0.22 |
| chr9_68906459_68906626 | 0.14 |
Rora |
RAR-related orphan receptor alpha |
251239 |
0.02 |
| chr2_144256463_144256614 | 0.14 |
Snx5 |
sorting nexin 5 |
1135 |
0.31 |
| chr17_80728516_80728734 | 0.14 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
140 |
0.84 |
| chr5_97876322_97876543 | 0.14 |
Antxr2 |
anthrax toxin receptor 2 |
119663 |
0.05 |
| chr17_46160193_46160368 | 0.14 |
Gtpbp2 |
GTP binding protein 2 |
752 |
0.48 |
| chr13_21778741_21779257 | 0.14 |
H1f5 |
H1.5 linker histone, cluster member |
1626 |
0.11 |
| chrX_74837196_74837401 | 0.14 |
Gm5936 |
predicted gene 5936 |
102 |
0.92 |
| chr11_16806184_16806397 | 0.14 |
Egfros |
epidermal growth factor receptor, opposite strand |
24412 |
0.18 |
| chr19_43803990_43804141 | 0.14 |
Abcc2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
6079 |
0.16 |
| chr4_151045208_151045385 | 0.14 |
Per3 |
period circadian clock 3 |
631 |
0.67 |
| chr12_79830687_79831018 | 0.14 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
93881 |
0.08 |
| chr12_103947018_103947616 | 0.13 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
9581 |
0.11 |
| chrX_11273388_11273552 | 0.13 |
H2al1a |
H2A histone family member L1A |
25787 |
0.09 |
| chr7_115857826_115857977 | 0.13 |
Sox6 |
SRY (sex determining region Y)-box 6 |
1951 |
0.46 |
| chr1_87764802_87764962 | 0.13 |
Atg16l1 |
autophagy related 16-like 1 (S. cerevisiae) |
2019 |
0.23 |
| chr2_176830816_176830976 | 0.13 |
Gm14408 |
predicted gene 14408 |
244 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.2 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.1 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.0 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |