Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
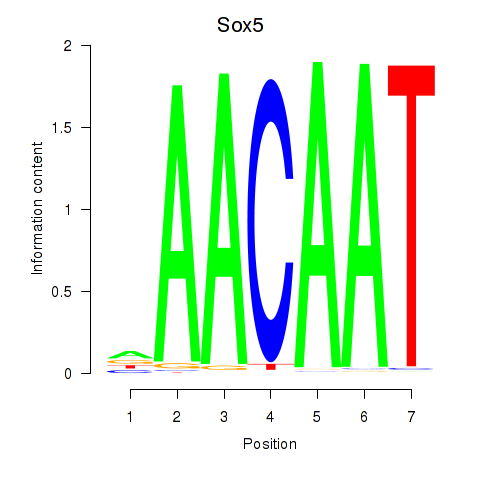
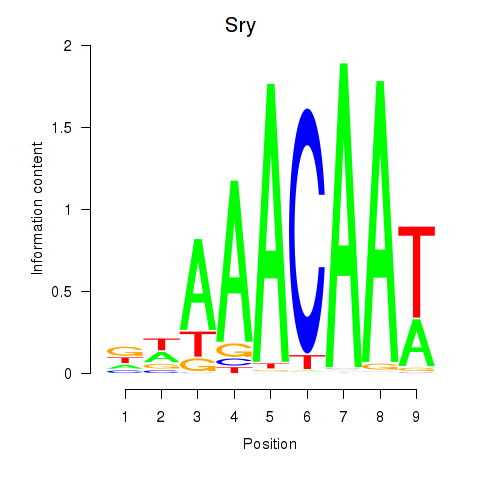
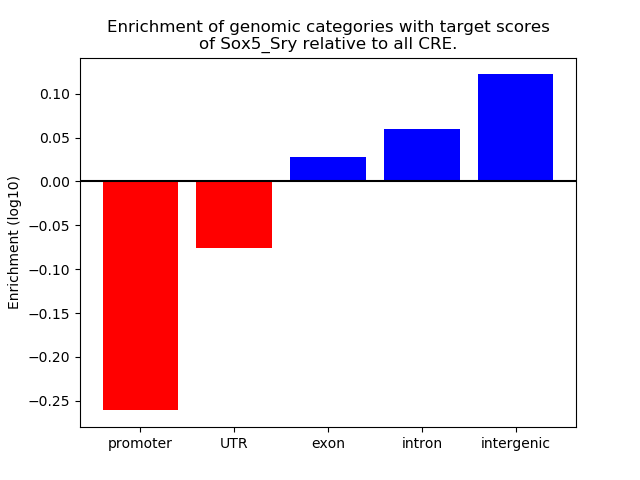
Results for Sox5_Sry
Z-value: 0.46
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSMUSG00000041540.10 | SRY (sex determining region Y)-box 5 |
|
Sry
|
ENSMUSG00000069036.3 | sex determining region of Chr Y |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_144155774_144155925 | Sox5 | 48208 | 0.183274 | -0.91 | 1.2e-02 | Click! |
| chr6_144204039_144204246 | Sox5 | 85 | 0.985389 | -0.81 | 5.1e-02 | Click! |
| chr6_144753669_144753921 | Sox5 | 28124 | 0.135933 | -0.80 | 5.6e-02 | Click! |
| chr6_144210184_144210365 | Sox5 | 706 | 0.810293 | -0.80 | 5.7e-02 | Click! |
| chr6_144209110_144209295 | Sox5 | 250 | 0.959818 | -0.78 | 6.5e-02 | Click! |
Activity of the Sox5_Sry motif across conditions
Conditions sorted by the z-value of the Sox5_Sry motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_116510337_116510525 | 0.17 |
Rtca |
RNA 3'-terminal phosphate cyclase |
2223 |
0.16 |
| chr11_16704048_16704199 | 0.16 |
Gm25698 |
predicted gene, 25698 |
28588 |
0.15 |
| chr7_140772355_140773139 | 0.15 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
8266 |
0.09 |
| chr3_69094579_69094744 | 0.14 |
Gm22009 |
predicted gene, 22009 |
9214 |
0.13 |
| chr19_40156899_40157166 | 0.14 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
30254 |
0.13 |
| chr15_59391938_59392317 | 0.14 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
17840 |
0.18 |
| chr1_92683263_92683419 | 0.14 |
Gm29483 |
predicted gene 29483 |
33861 |
0.09 |
| chr1_74807738_74808135 | 0.14 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
14573 |
0.11 |
| chr2_147988721_147988917 | 0.13 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
12634 |
0.21 |
| chr9_74872523_74872674 | 0.13 |
Onecut1 |
one cut domain, family member 1 |
6114 |
0.16 |
| chr15_102116938_102117135 | 0.13 |
Tns2 |
tensin 2 |
1897 |
0.2 |
| chr16_24121120_24121271 | 0.12 |
Gm31583 |
predicted gene, 31583 |
31106 |
0.14 |
| chr4_114958381_114958568 | 0.12 |
Cmpk1 |
cytidine monophosphate (UMP-CMP) kinase 1 |
28598 |
0.11 |
| chr12_104083020_104083260 | 0.12 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr12_37050724_37050882 | 0.11 |
Gm48617 |
predicted gene, 48617 |
1586 |
0.37 |
| chr7_134497342_134497526 | 0.11 |
D7Ertd443e |
DNA segment, Chr 7, ERATO Doi 443, expressed |
1550 |
0.55 |
| chr18_16662543_16662742 | 0.10 |
Cdh2 |
cadherin 2 |
7427 |
0.29 |
| chr15_26639292_26639449 | 0.10 |
9430068D22Rik |
RIKEN cDNA 9430068D22 gene |
5919 |
0.2 |
| chr11_16821774_16821942 | 0.10 |
Egfros |
epidermal growth factor receptor, opposite strand |
8844 |
0.22 |
| chr16_49049395_49049546 | 0.10 |
Myh15 |
myosin, heavy chain 15 |
8016 |
0.22 |
| chr1_88064051_88064407 | 0.10 |
AC087801.2 |
UDP glucuronosyltransferase 1 family, polypeptide A9 (Ugt1a9), pseudogene |
3105 |
0.1 |
| chr3_80042843_80043016 | 0.10 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
11963 |
0.22 |
| chr2_178471902_178472068 | 0.10 |
Cdh26 |
cadherin-like 26 |
11355 |
0.22 |
| chr14_66023636_66023794 | 0.10 |
Gulo |
gulonolactone (L-) oxidase |
14508 |
0.15 |
| chr3_25017765_25017942 | 0.09 |
Gm42774 |
predicted gene 42774 |
10148 |
0.29 |
| chr13_90739139_90739300 | 0.09 |
Gm44338 |
predicted gene, 44338 |
20893 |
0.2 |
| chr2_149993027_149993210 | 0.09 |
Gm14140 |
predicted gene 14140 |
118458 |
0.05 |
| chr11_49837892_49838056 | 0.09 |
Mapk9 |
mitogen-activated protein kinase 9 |
8777 |
0.16 |
| chr10_59537699_59537863 | 0.09 |
Gm10322 |
predicted gene 10322 |
78282 |
0.08 |
| chr10_20749070_20749231 | 0.09 |
Gm48651 |
predicted gene, 48651 |
6967 |
0.23 |
| chr3_10136245_10136401 | 0.09 |
Gm37308 |
predicted gene, 37308 |
46368 |
0.08 |
| chr2_84180208_84180377 | 0.09 |
Gm25972 |
predicted gene, 25972 |
30237 |
0.16 |
| chr1_88048809_88049389 | 0.08 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
6289 |
0.08 |
| chr13_44026422_44026628 | 0.08 |
Gm33489 |
predicted gene, 33489 |
8003 |
0.22 |
| chr2_11597723_11597874 | 0.08 |
Rbm17 |
RNA binding motif protein 17 |
5464 |
0.14 |
| chr1_71807937_71808302 | 0.08 |
Gm37217 |
predicted gene, 37217 |
38491 |
0.15 |
| chr18_43980831_43980982 | 0.08 |
Spink5 |
serine peptidase inhibitor, Kazal type 5 |
17671 |
0.17 |
| chr9_32656566_32656728 | 0.08 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
20399 |
0.15 |
| chr9_100488481_100488638 | 0.08 |
Il20rb |
interleukin 20 receptor beta |
1771 |
0.29 |
| chr2_22587699_22587875 | 0.08 |
Gm13341 |
predicted gene 13341 |
175 |
0.91 |
| chr14_7840036_7840206 | 0.08 |
Flnb |
filamin, beta |
22164 |
0.15 |
| chr17_5324481_5324860 | 0.08 |
Gm29050 |
predicted gene 29050 |
64093 |
0.11 |
| chr6_77098640_77098806 | 0.08 |
Ctnna2 |
catenin (cadherin associated protein), alpha 2 |
45220 |
0.19 |
| chr6_101160855_101161061 | 0.08 |
Gm19726 |
predicted gene, 19726 |
14479 |
0.15 |
| chr2_83740744_83740901 | 0.08 |
Dnmt3l-ps1 |
DNA methyltransferase 3-like, pseudogene 1 |
4914 |
0.17 |
| chr13_110398929_110399098 | 0.08 |
Plk2 |
polo like kinase 2 |
1216 |
0.56 |
| chr5_125518895_125519046 | 0.08 |
Aacs |
acetoacetyl-CoA synthetase |
3727 |
0.19 |
| chr4_70531154_70531323 | 0.08 |
Megf9 |
multiple EGF-like-domains 9 |
3690 |
0.37 |
| chr13_63298050_63298241 | 0.08 |
Aopep |
aminopeptidase O |
648 |
0.44 |
| chr3_121982496_121982688 | 0.08 |
Arhgap29 |
Rho GTPase activating protein 29 |
1042 |
0.54 |
| chr4_80003665_80003823 | 0.08 |
Gm11408 |
predicted gene 11408 |
170 |
0.59 |
| chr15_6248496_6248831 | 0.08 |
Dab2 |
disabled 2, mitogen-responsive phosphoprotein |
51125 |
0.13 |
| chr4_55750022_55750212 | 0.08 |
Gm12506 |
predicted gene 12506 |
144968 |
0.04 |
| chr9_53288790_53288968 | 0.08 |
Exph5 |
exophilin 5 |
12791 |
0.18 |
| chr7_140767825_140767976 | 0.07 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
3419 |
0.12 |
| chr16_91411218_91411402 | 0.07 |
Il10rb |
interleukin 10 receptor, beta |
4866 |
0.12 |
| chrX_116629798_116629984 | 0.07 |
H2ab2 |
H2A.B variant histone 2 |
51287 |
0.17 |
| chr16_23326741_23326947 | 0.07 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
36374 |
0.13 |
| chr17_31079236_31079399 | 0.07 |
Gm25447 |
predicted gene, 25447 |
19713 |
0.12 |
| chr13_80886812_80887097 | 0.07 |
Arrdc3 |
arrestin domain containing 3 |
859 |
0.49 |
| chr1_74027353_74027507 | 0.07 |
Tns1 |
tensin 1 |
9703 |
0.23 |
| chr2_128719461_128719612 | 0.07 |
Mertk |
MER proto-oncogene tyrosine kinase |
20580 |
0.14 |
| chr9_85369091_85369242 | 0.07 |
Gm48832 |
predicted gene, 48832 |
21101 |
0.14 |
| chr10_37711570_37711751 | 0.07 |
Gm24710 |
predicted gene, 24710 |
220020 |
0.02 |
| chr2_46365879_46366164 | 0.07 |
Gm13469 |
predicted gene 13469 |
40548 |
0.18 |
| chr1_172525495_172525646 | 0.07 |
Cfap45 |
cilia and flagella associated protein 45 |
479 |
0.67 |
| chr1_70811018_70811204 | 0.07 |
Vwc2l |
von Willebrand factor C domain-containing protein 2-like |
85188 |
0.1 |
| chr5_74647970_74648138 | 0.07 |
Lnx1 |
ligand of numb-protein X 1 |
24533 |
0.16 |
| chr10_19904541_19904692 | 0.07 |
Pex7 |
peroxisomal biogenesis factor 7 |
3031 |
0.24 |
| chr5_92858921_92859073 | 0.07 |
Shroom3 |
shroom family member 3 |
2526 |
0.33 |
| chr2_163586943_163587113 | 0.07 |
Ttpal |
tocopherol (alpha) transfer protein-like |
15286 |
0.13 |
| chr5_17836112_17836269 | 0.07 |
Cd36 |
CD36 molecule |
473 |
0.89 |
| chr4_86047006_86047165 | 0.07 |
Gm25811 |
predicted gene, 25811 |
3591 |
0.3 |
| chr4_59292456_59292607 | 0.07 |
Susd1 |
sushi domain containing 1 |
23435 |
0.17 |
| chr18_60748821_60749236 | 0.07 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
378 |
0.77 |
| chr3_39220363_39220530 | 0.07 |
Gm43008 |
predicted gene 43008 |
2600 |
0.39 |
| chr8_11148911_11149083 | 0.07 |
Gm44717 |
predicted gene 44717 |
249 |
0.91 |
| chr10_80917355_80917522 | 0.07 |
Lmnb2 |
lamin B2 |
413 |
0.7 |
| chr6_91424605_91424772 | 0.07 |
Gm4575 |
predicted gene 4575 |
4195 |
0.12 |
| chr18_78260055_78260206 | 0.07 |
Slc14a2 |
solute carrier family 14 (urea transporter), member 2 |
51036 |
0.15 |
| chr11_107402263_107402420 | 0.07 |
Gm11713 |
predicted gene 11713 |
8049 |
0.14 |
| chr11_107276986_107277168 | 0.07 |
Gm11719 |
predicted gene 11719 |
53631 |
0.09 |
| chr12_54671337_54671527 | 0.07 |
Gm24971 |
predicted gene, 24971 |
4646 |
0.08 |
| chr19_27672733_27673067 | 0.07 |
Gm24782 |
predicted gene, 24782 |
31107 |
0.23 |
| chr1_60716026_60716187 | 0.07 |
Cd28 |
CD28 antigen |
694 |
0.59 |
| chr13_90799244_90799545 | 0.07 |
Gm47521 |
predicted gene, 47521 |
21311 |
0.19 |
| chr1_174742793_174742954 | 0.07 |
Fmn2 |
formin 2 |
28547 |
0.24 |
| chr14_31101945_31102096 | 0.07 |
Pbrm1 |
polybromo 1 |
17816 |
0.09 |
| chr19_12613775_12613971 | 0.07 |
Gm4952 |
predicted gene 4952 |
13857 |
0.09 |
| chr6_31148380_31148897 | 0.07 |
Gm37728 |
predicted gene, 37728 |
1219 |
0.36 |
| chr10_126001088_126001318 | 0.06 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
10428 |
0.26 |
| chr17_73786615_73786813 | 0.06 |
Ehd3 |
EH-domain containing 3 |
17420 |
0.2 |
| chr17_27480362_27480514 | 0.06 |
Grm4 |
glutamate receptor, metabotropic 4 |
7122 |
0.11 |
| chr10_87030552_87030730 | 0.06 |
Stab2 |
stabilin 2 |
22616 |
0.14 |
| chr1_134079839_134079990 | 0.06 |
Btg2 |
BTG anti-proliferation factor 2 |
794 |
0.53 |
| chr1_170972984_170973148 | 0.06 |
Gm38204 |
predicted gene, 38204 |
307 |
0.79 |
| chr6_141348213_141348590 | 0.06 |
Gm10400 |
predicted gene 10400 |
7848 |
0.26 |
| chr13_94037879_94038063 | 0.06 |
Cycs-ps3 |
cytochrome c, pseudogene 3 |
15872 |
0.17 |
| chr15_25531182_25531388 | 0.06 |
Gm36642 |
predicted gene, 36642 |
61806 |
0.1 |
| chr8_109993075_109993269 | 0.06 |
Tat |
tyrosine aminotransferase |
2666 |
0.18 |
| chr19_36803505_36803664 | 0.06 |
Gm50112 |
predicted gene, 50112 |
15064 |
0.19 |
| chr6_35253418_35253775 | 0.06 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
864 |
0.54 |
| chr13_107798210_107798399 | 0.06 |
Gm22118 |
predicted gene, 22118 |
30976 |
0.19 |
| chr10_84581307_84581543 | 0.06 |
Tcp11l2 |
t-complex 11 (mouse) like 2 |
4528 |
0.15 |
| chr9_70011238_70011422 | 0.06 |
Gtf2a2 |
general transcription factor II A, 2 |
1220 |
0.39 |
| chr6_113038436_113038611 | 0.06 |
Gm23244 |
predicted gene, 23244 |
2589 |
0.16 |
| chr1_60833760_60833934 | 0.06 |
Gm11581 |
predicted gene 11581 |
24731 |
0.11 |
| chr1_88044946_88045337 | 0.06 |
AC087801.1 |
UDP glycosyltransferase 1 family (Ytg1) pseudogene |
7541 |
0.08 |
| chrX_20561959_20562110 | 0.06 |
Rgn |
regucalcin |
12247 |
0.13 |
| chr1_72255173_72255368 | 0.06 |
Gm25939 |
predicted gene, 25939 |
262 |
0.86 |
| chr2_168102539_168102698 | 0.06 |
AL831766.1 |
breast carcinoma amplified sequence 4 (BCAS4) pseudogene |
12259 |
0.13 |
| chr3_99187907_99188075 | 0.06 |
Gm18982 |
predicted gene, 18982 |
6710 |
0.13 |
| chr12_58297089_58297242 | 0.06 |
Clec14a |
C-type lectin domain family 14, member a |
27875 |
0.24 |
| chr17_80677489_80677679 | 0.06 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
26434 |
0.19 |
| chr1_57391527_57391708 | 0.06 |
Tyw5 |
tRNA-yW synthesizing protein 5 |
123 |
0.95 |
| chr11_107373329_107373545 | 0.06 |
Gm11712 |
predicted gene 11712 |
13546 |
0.13 |
| chr8_93233702_93233868 | 0.06 |
Ces1e |
carboxylesterase 1E |
4166 |
0.16 |
| chr8_106882742_106882961 | 0.06 |
Chtf8 |
CTF8, chromosome transmission fidelity factor 8 |
10364 |
0.12 |
| chr7_126760516_126760683 | 0.06 |
Mapk3 |
mitogen-activated protein kinase 3 |
2 |
0.92 |
| chr5_3488489_3488661 | 0.06 |
Gm17590 |
predicted gene, 17590 |
7702 |
0.13 |
| chrX_120279500_120279651 | 0.06 |
Pcdh11x |
protocadherin 11 X-linked |
10684 |
0.2 |
| chr3_121346684_121346853 | 0.06 |
Gm5711 |
predicted gene 5711 |
33911 |
0.12 |
| chr5_130200757_130200925 | 0.06 |
Rabgef1 |
RAB guanine nucleotide exchange factor (GEF) 1 |
6212 |
0.1 |
| chr3_63992313_63992468 | 0.06 |
Gmps |
guanine monophosphate synthetase |
16284 |
0.13 |
| chr1_64308159_64308318 | 0.06 |
Gm28981 |
predicted gene 28981 |
5195 |
0.28 |
| chr5_43788120_43788279 | 0.06 |
Gm43181 |
predicted gene 43181 |
2082 |
0.19 |
| chr14_65357765_65357928 | 0.06 |
Zfp395 |
zinc finger protein 395 |
543 |
0.68 |
| chr1_31046483_31046963 | 0.06 |
Gm28644 |
predicted gene 28644 |
4804 |
0.19 |
| chr4_53280466_53280664 | 0.06 |
AI427809 |
expressed sequence AI427809 |
10333 |
0.18 |
| chr15_73506117_73506298 | 0.06 |
Dennd3 |
DENN/MADD domain containing 3 |
6353 |
0.18 |
| chr16_46131552_46131725 | 0.06 |
Cd96 |
CD96 antigen |
11387 |
0.19 |
| chr8_69063559_69063737 | 0.06 |
Slc18a1 |
solute carrier family 18 (vesicular monoamine), member 1 |
11860 |
0.15 |
| chr4_49535372_49535819 | 0.06 |
Aldob |
aldolase B, fructose-bisphosphate |
3271 |
0.17 |
| chr14_86309035_86309286 | 0.06 |
Gm23926 |
predicted gene, 23926 |
2218 |
0.22 |
| chr5_112843602_112843780 | 0.06 |
Myo18b |
myosin XVIIIb |
27895 |
0.17 |
| chr10_121316471_121316644 | 0.06 |
Tbc1d30 |
TBC1 domain family, member 30 |
5368 |
0.16 |
| chr12_51709142_51709293 | 0.06 |
Strn3 |
striatin, calmodulin binding protein 3 |
17320 |
0.16 |
| chr4_49542856_49543140 | 0.06 |
Aldob |
aldolase B, fructose-bisphosphate |
3112 |
0.18 |
| chr8_76907164_76907353 | 0.06 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
1014 |
0.57 |
| chr4_137772777_137772941 | 0.06 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
3039 |
0.27 |
| chr4_81591457_81591882 | 0.06 |
Gm11411 |
predicted gene 11411 |
104772 |
0.07 |
| chr5_147894276_147894456 | 0.05 |
Slc46a3 |
solute carrier family 46, member 3 |
449 |
0.8 |
| chr19_10664062_10664213 | 0.05 |
Vwce |
von Willebrand factor C and EGF domains |
6148 |
0.1 |
| chr5_90457077_90457513 | 0.05 |
Alb |
albumin |
3602 |
0.2 |
| chr19_8097849_8098000 | 0.05 |
Slc22a28 |
solute carrier family 22, member 28 |
34058 |
0.16 |
| chr3_89143426_89143577 | 0.05 |
Pklr |
pyruvate kinase liver and red blood cell |
6878 |
0.07 |
| chr7_140719940_140720091 | 0.05 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
1267 |
0.28 |
| chr16_95432244_95432403 | 0.05 |
Erg |
ETS transcription factor |
26922 |
0.22 |
| chr15_10375239_10375408 | 0.05 |
Agxt2 |
alanine-glyoxylate aminotransferase 2 |
16744 |
0.19 |
| chr11_16845424_16845700 | 0.05 |
Egfros |
epidermal growth factor receptor, opposite strand |
14860 |
0.2 |
| chr16_95459306_95459486 | 0.05 |
Erg |
ETS transcription factor |
48 |
0.99 |
| chr10_93282645_93282796 | 0.05 |
Elk3 |
ELK3, member of ETS oncogene family |
28091 |
0.14 |
| chr13_114392779_114392930 | 0.05 |
Ndufs4 |
NADH:ubiquinone oxidoreductase core subunit S4 |
4596 |
0.21 |
| chr5_139384486_139384856 | 0.05 |
Gpr146 |
G protein-coupled receptor 146 |
4090 |
0.13 |
| chr14_37027574_37027733 | 0.05 |
Gm47885 |
predicted gene, 47885 |
3119 |
0.2 |
| chr8_69112104_69112255 | 0.05 |
Atp6v1b2 |
ATPase, H+ transporting, lysosomal V1 subunit B2 |
9051 |
0.16 |
| chr15_75596455_75596620 | 0.05 |
Gpihbp1 |
GPI-anchored HDL-binding protein 1 |
91 |
0.95 |
| chr17_45567292_45567749 | 0.05 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
2216 |
0.14 |
| chr3_114294859_114295010 | 0.05 |
Col11a1 |
collagen, type XI, alpha 1 |
129656 |
0.06 |
| chr17_31474403_31474582 | 0.05 |
Pde9a |
phosphodiesterase 9A |
3440 |
0.12 |
| chr4_150576385_150576576 | 0.05 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
6823 |
0.21 |
| chr6_5473962_5474316 | 0.05 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
22122 |
0.24 |
| chr5_107120720_107120871 | 0.05 |
Tgfbr3 |
transforming growth factor, beta receptor III |
12128 |
0.16 |
| chr8_126847749_126848310 | 0.05 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
8796 |
0.2 |
| chr13_35016383_35016558 | 0.05 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
10624 |
0.12 |
| chr4_6539560_6539735 | 0.05 |
Gm11801 |
predicted gene 11801 |
86 |
0.98 |
| chr6_149359539_149359690 | 0.05 |
Gm15784 |
predicted gene 15784 |
2072 |
0.23 |
| chr7_73478544_73478710 | 0.05 |
Chd2 |
chromodomain helicase DNA binding protein 2 |
201 |
0.91 |
| chr2_103870054_103870389 | 0.05 |
Gm13876 |
predicted gene 13876 |
18103 |
0.09 |
| chr3_94512708_94512862 | 0.05 |
Snx27 |
sorting nexin family member 27 |
6306 |
0.09 |
| chr5_100368600_100368758 | 0.05 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
4409 |
0.19 |
| chr2_155539663_155540189 | 0.05 |
Mipep-ps |
mitochondrial intermediate peptidase, pseudogene |
2721 |
0.13 |
| chr13_25791444_25791595 | 0.05 |
Gm11350 |
predicted gene 11350 |
110376 |
0.07 |
| chr3_18877716_18877898 | 0.05 |
Gm30341 |
predicted gene, 30341 |
124730 |
0.06 |
| chr8_115089778_115090403 | 0.05 |
Gm22556 |
predicted gene, 22556 |
37177 |
0.23 |
| chr1_173599258_173599409 | 0.05 |
Ifi213 |
interferon activated gene 213 |
59 |
0.97 |
| chr2_96253373_96253545 | 0.05 |
Lrrc4c |
leucine rich repeat containing 4C |
64710 |
0.14 |
| chr1_173491252_173491410 | 0.05 |
Ifi206 |
interferon activated gene 206 |
290 |
0.89 |
| chr6_128522025_128522231 | 0.05 |
Pzp |
PZP, alpha-2-macroglobulin like |
4575 |
0.11 |
| chr1_136441030_136441213 | 0.05 |
Ddx59 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
8719 |
0.15 |
| chr1_54850108_54850451 | 0.05 |
Ankrd44 |
ankyrin repeat domain 44 |
34432 |
0.16 |
| chr2_102694561_102694718 | 0.05 |
Slc1a2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
11717 |
0.23 |
| chr13_24945328_24945479 | 0.05 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
2251 |
0.21 |
| chr4_144977157_144977352 | 0.05 |
Vps13d |
vacuolar protein sorting 13D |
6707 |
0.21 |
| chr16_91434651_91434809 | 0.05 |
Gm46562 |
predicted gene, 46562 |
23691 |
0.08 |
| chr15_62529162_62529323 | 0.05 |
Gm41333 |
predicted gene, 41333 |
38733 |
0.21 |
| chr18_5091846_5091997 | 0.05 |
Svil |
supervillin |
2460 |
0.4 |
| chr13_44890716_44890885 | 0.05 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
50017 |
0.13 |
| chr18_39357013_39357188 | 0.05 |
Arhgap26 |
Rho GTPase activating protein 26 |
5483 |
0.2 |
| chr12_41081814_41082149 | 0.05 |
Immp2l |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
13669 |
0.21 |
| chr10_26828824_26829011 | 0.05 |
Arhgap18 |
Rho GTPase activating protein 18 |
6315 |
0.25 |
| chr15_43702286_43702631 | 0.05 |
Gm2140 |
predicted gene 2140 |
106003 |
0.07 |
| chr17_46031864_46032231 | 0.05 |
Vegfa |
vascular endothelial growth factor A |
232 |
0.92 |
| chr18_10413650_10413816 | 0.05 |
Greb1l |
growth regulation by estrogen in breast cancer-like |
44492 |
0.12 |
| chr11_50175754_50175938 | 0.05 |
Mrnip |
MRN complex interacting protein |
939 |
0.42 |
| chrX_84312323_84312721 | 0.05 |
Dmd |
dystrophin, muscular dystrophy |
235873 |
0.02 |
| chr5_100371529_100371680 | 0.05 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
1880 |
0.3 |
| chr15_55245352_55245503 | 0.05 |
Gm26296 |
predicted gene, 26296 |
17176 |
0.21 |
| chr4_141161528_141161683 | 0.05 |
Fbxo42 |
F-box protein 42 |
13683 |
0.11 |
| chr13_72293404_72293565 | 0.05 |
Gm4052 |
predicted gene 4052 |
56737 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |