Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
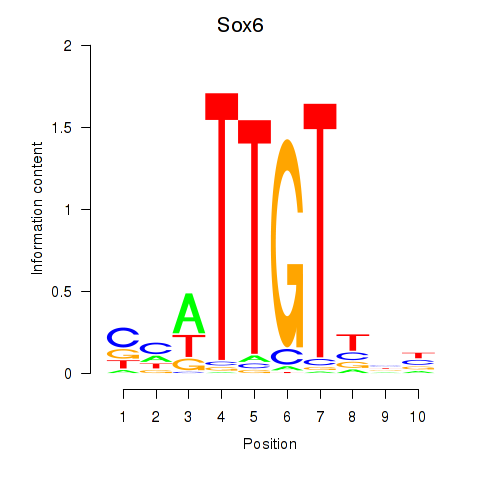
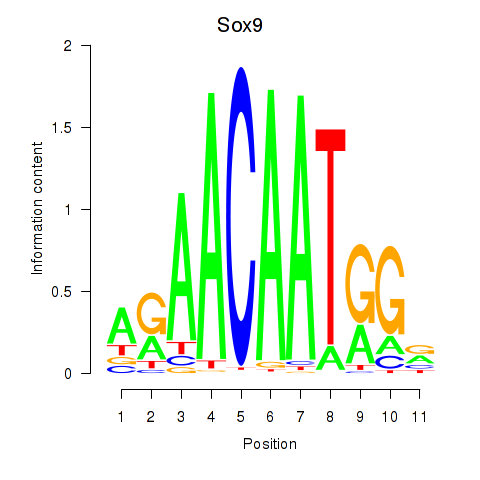
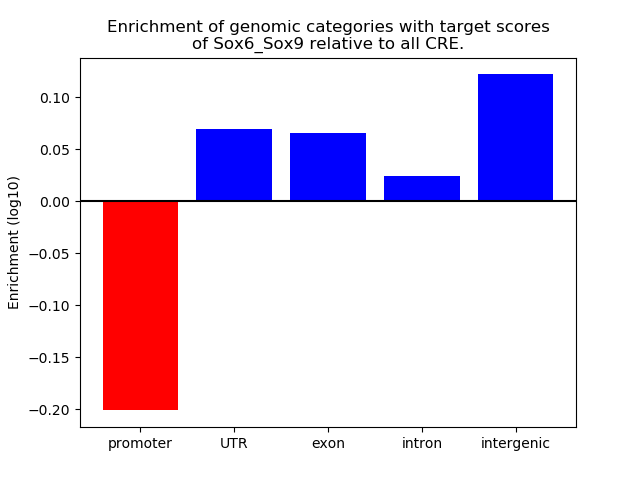
Results for Sox6_Sox9
Z-value: 1.72
Transcription factors associated with Sox6_Sox9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox6
|
ENSMUSG00000051910.7 | SRY (sex determining region Y)-box 6 |
|
Sox9
|
ENSMUSG00000000567.5 | SRY (sex determining region Y)-box 9 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_115576330_115576481 | Sox6 | 21491 | 0.280207 | -0.92 | 1.0e-02 | Click! |
| chr7_115731752_115731939 | Sox6 | 69480 | 0.131585 | -0.89 | 1.7e-02 | Click! |
| chr7_115644077_115644258 | Sox6 | 18198 | 0.288844 | -0.87 | 2.5e-02 | Click! |
| chr7_115661755_115661917 | Sox6 | 529 | 0.877130 | -0.82 | 4.5e-02 | Click! |
| chr7_115857826_115857977 | Sox6 | 1951 | 0.461715 | -0.82 | 4.6e-02 | Click! |
| chr11_112788056_112788236 | Sox9 | 5922 | 0.187637 | 0.88 | 2.2e-02 | Click! |
| chr11_112783351_112783652 | Sox9 | 1277 | 0.363013 | 0.74 | 9.1e-02 | Click! |
| chr11_112788385_112788735 | Sox9 | 6336 | 0.185303 | 0.71 | 1.1e-01 | Click! |
| chr11_112782073_112782233 | Sox9 | 71 | 0.650859 | 0.56 | 2.5e-01 | Click! |
Activity of the Sox6_Sox9 motif across conditions
Conditions sorted by the z-value of the Sox6_Sox9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
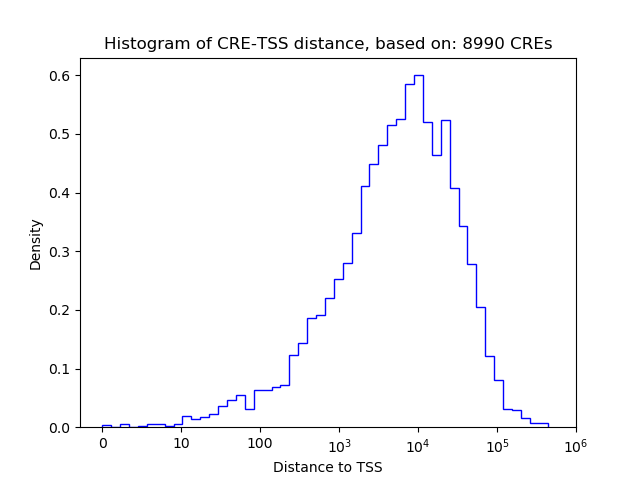
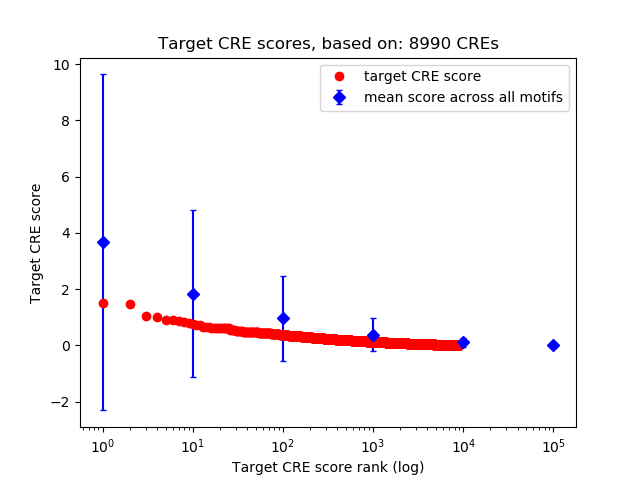
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_93624884_93625040 | 1.49 |
Gm15622 |
predicted gene 15622 |
420 |
0.81 |
| chr5_8968601_8968752 | 1.46 |
Gm15610 |
predicted gene 15610 |
3272 |
0.16 |
| chr10_85161408_85161709 | 1.05 |
Cry1 |
cryptochrome 1 (photolyase-like) |
10202 |
0.18 |
| chr4_60659551_60660018 | 0.99 |
Mup11 |
major urinary protein 11 |
46 |
0.97 |
| chr4_60418315_60418583 | 0.91 |
Mup9 |
major urinary protein 9 |
2135 |
0.24 |
| chr9_104461716_104461890 | 0.90 |
Gm25904 |
predicted gene, 25904 |
19707 |
0.19 |
| chr4_60498312_60498676 | 0.88 |
Mup1 |
major urinary protein 1 |
838 |
0.49 |
| chr4_60066787_60066938 | 0.84 |
Mup7 |
major urinary protein 7 |
3549 |
0.22 |
| chr13_93625562_93625714 | 0.80 |
Gm15622 |
predicted gene 15622 |
256 |
0.91 |
| chr5_8964466_8964675 | 0.76 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
6018 |
0.13 |
| chr4_60218940_60219116 | 0.73 |
Mup8 |
major urinary protein 8 |
3552 |
0.23 |
| chr11_28906435_28906594 | 0.72 |
Efemp1 |
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
9153 |
0.23 |
| chr6_129215204_129215355 | 0.66 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
18695 |
0.1 |
| chr3_85858327_85858483 | 0.66 |
Gm37240 |
predicted gene, 37240 |
28963 |
0.1 |
| chr3_83023876_83024027 | 0.63 |
Fga |
fibrinogen alpha chain |
2125 |
0.24 |
| chr18_20974221_20974372 | 0.63 |
Rnf125 |
ring finger protein 125 |
12824 |
0.2 |
| chr10_8122314_8122465 | 0.62 |
Gm30906 |
predicted gene, 30906 |
53742 |
0.15 |
| chr5_9047464_9048021 | 0.62 |
Gm40264 |
predicted gene, 40264 |
12618 |
0.15 |
| chr9_65334133_65334359 | 0.61 |
Gm39363 |
predicted gene, 39363 |
1726 |
0.18 |
| chr10_84418574_84418728 | 0.61 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
21946 |
0.15 |
| chr5_8961694_8961940 | 0.61 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
3265 |
0.17 |
| chr19_41334643_41334794 | 0.61 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
50378 |
0.14 |
| chr8_11308621_11308772 | 0.60 |
Col4a1 |
collagen, type IV, alpha 1 |
3993 |
0.18 |
| chr1_58170640_58170791 | 0.60 |
Aox3 |
aldehyde oxidase 3 |
1091 |
0.5 |
| chr14_37027574_37027733 | 0.60 |
Gm47885 |
predicted gene, 47885 |
3119 |
0.2 |
| chr7_101479062_101479229 | 0.56 |
Pde2a |
phosphodiesterase 2A, cGMP-stimulated |
2140 |
0.23 |
| chr2_115520314_115520525 | 0.54 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
7325 |
0.25 |
| chr12_8643417_8643595 | 0.54 |
Pum2 |
pumilio RNA-binding family member 2 |
30628 |
0.17 |
| chr6_54842890_54843041 | 0.54 |
Znrf2 |
zinc and ring finger 2 |
25517 |
0.18 |
| chr19_3669127_3669720 | 0.51 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
17133 |
0.12 |
| chr18_20984644_20984795 | 0.51 |
Rnf138 |
ring finger protein 138 |
16622 |
0.19 |
| chr11_59965854_59966027 | 0.50 |
Rasd1 |
RAS, dexamethasone-induced 1 |
996 |
0.42 |
| chr8_27233050_27233201 | 0.50 |
Gm45470 |
predicted gene 45470 |
747 |
0.48 |
| chr14_40949121_40949299 | 0.50 |
Tspan14 |
tetraspanin 14 |
14889 |
0.18 |
| chr10_12816564_12816741 | 0.50 |
Utrn |
utrophin |
3037 |
0.26 |
| chrX_169863890_169864041 | 0.49 |
Mid1 |
midline 1 |
15657 |
0.24 |
| chr11_112217336_112217487 | 0.49 |
Gm11680 |
predicted gene 11680 |
109973 |
0.07 |
| chr1_88975960_88976113 | 0.49 |
1700067G17Rik |
RIKEN cDNA 1700067G17 gene |
40077 |
0.14 |
| chr5_117361300_117361465 | 0.48 |
Wsb2 |
WD repeat and SOCS box-containing 2 |
360 |
0.77 |
| chr1_180219918_180220099 | 0.48 |
Gm37336 |
predicted gene, 37336 |
7607 |
0.14 |
| chr14_30908290_30908855 | 0.48 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
5557 |
0.12 |
| chr8_36723231_36723557 | 0.48 |
Dlc1 |
deleted in liver cancer 1 |
9660 |
0.29 |
| chr3_18064935_18065086 | 0.48 |
Gm23726 |
predicted gene, 23726 |
4316 |
0.23 |
| chr13_93618232_93618425 | 0.48 |
Gm15622 |
predicted gene 15622 |
7054 |
0.17 |
| chr10_93506452_93506730 | 0.47 |
Hal |
histidine ammonia lyase |
5466 |
0.15 |
| chr13_36466242_36466419 | 0.47 |
Gm48763 |
predicted gene, 48763 |
7050 |
0.24 |
| chr11_72281229_72281388 | 0.47 |
n-R5s70 |
nuclear encoded rRNA 5S 70 |
292 |
0.83 |
| chr9_105759000_105759170 | 0.46 |
Gm38314 |
predicted gene, 38314 |
31300 |
0.16 |
| chr6_72633856_72634007 | 0.46 |
Gm15401 |
predicted gene 15401 |
2322 |
0.13 |
| chr9_65333970_65334121 | 0.46 |
Gm39363 |
predicted gene, 39363 |
1525 |
0.2 |
| chr17_75432717_75432868 | 0.46 |
Rasgrp3 |
RAS, guanyl releasing protein 3 |
3104 |
0.34 |
| chrX_13275902_13276062 | 0.46 |
Ddx3x |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked |
4988 |
0.14 |
| chr5_28003991_28004186 | 0.46 |
Gm4865 |
predicted gene 4865 |
2782 |
0.24 |
| chr14_8248346_8248505 | 0.45 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
1995 |
0.33 |
| chr10_85193452_85193650 | 0.45 |
Cry1 |
cryptochrome 1 (photolyase-like) |
8487 |
0.2 |
| chr18_20931708_20931938 | 0.45 |
Rnf125 |
ring finger protein 125 |
12802 |
0.2 |
| chr13_43500025_43500247 | 0.45 |
Gm47683 |
predicted gene, 47683 |
31 |
0.97 |
| chr15_10960635_10960786 | 0.45 |
C1qtnf3 |
C1q and tumor necrosis factor related protein 3 |
8353 |
0.15 |
| chr18_46725311_46725732 | 0.45 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
2508 |
0.19 |
| chr2_12382524_12382675 | 0.44 |
Mindy3 |
MINDY lysine 48 deubiquitinase 3 |
4182 |
0.17 |
| chr5_139938836_139939133 | 0.44 |
Gm43704 |
predicted gene 43704 |
24807 |
0.14 |
| chr3_133765600_133766533 | 0.44 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr14_22587357_22587508 | 0.44 |
Lrmda |
leucine rich melanocyte differentiation associated |
9081 |
0.23 |
| chr17_49349587_49349761 | 0.44 |
Gm18067 |
predicted gene, 18067 |
18178 |
0.18 |
| chr19_33098344_33098549 | 0.43 |
Gm29946 |
predicted gene, 29946 |
24201 |
0.18 |
| chr10_93497397_93497600 | 0.43 |
Hal |
histidine ammonia lyase |
1093 |
0.43 |
| chr10_45017413_45017609 | 0.43 |
Gm40639 |
predicted gene, 40639 |
6791 |
0.17 |
| chr19_36623611_36623788 | 0.43 |
Hectd2os |
Hectd2, opposite strand |
2305 |
0.33 |
| chr15_27564431_27564740 | 0.43 |
Ank |
progressive ankylosis |
6862 |
0.17 |
| chr5_150227882_150228033 | 0.42 |
Gm36378 |
predicted gene, 36378 |
15906 |
0.19 |
| chr16_31165540_31165735 | 0.42 |
Gm18237 |
predicted gene, 18237 |
7767 |
0.18 |
| chr2_3683170_3683338 | 0.42 |
Gm13183 |
predicted gene 13183 |
2664 |
0.22 |
| chr10_8133800_8133951 | 0.42 |
Gm30906 |
predicted gene, 30906 |
42256 |
0.18 |
| chr5_145854968_145855207 | 0.42 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
21604 |
0.14 |
| chr4_126097782_126097933 | 0.42 |
Lsm10 |
U7 snRNP-specific Sm-like protein LSM10 |
1197 |
0.32 |
| chr12_21167471_21167660 | 0.42 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
55611 |
0.1 |
| chr16_48767631_48767807 | 0.41 |
Trat1 |
T cell receptor associated transmembrane adaptor 1 |
4237 |
0.22 |
| chr10_8007720_8007891 | 0.41 |
Gm48614 |
predicted gene, 48614 |
13487 |
0.22 |
| chr10_59972679_59972846 | 0.41 |
Anapc16 |
anaphase promoting complex subunit 16 |
17621 |
0.15 |
| chr4_117682570_117682721 | 0.41 |
Dmap1 |
DNA methyltransferase 1-associated protein 1 |
372 |
0.82 |
| chr15_7088669_7089236 | 0.41 |
Lifr |
LIF receptor alpha |
1662 |
0.5 |
| chr13_114466635_114466797 | 0.41 |
Fst |
follistatin |
7765 |
0.16 |
| chr13_101895557_101895722 | 0.41 |
Gm17832 |
predicted gene, 17832 |
24781 |
0.19 |
| chr4_135962366_135962540 | 0.41 |
Gale |
galactose-4-epimerase, UDP |
1274 |
0.24 |
| chr19_30096271_30096642 | 0.40 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
4495 |
0.24 |
| chr8_120419577_120419740 | 0.40 |
Gm22715 |
predicted gene, 22715 |
23891 |
0.17 |
| chr11_96934737_96934922 | 0.40 |
Prr15l |
proline rich 15-like |
3442 |
0.11 |
| chr8_70224924_70225098 | 0.39 |
Armc6 |
armadillo repeat containing 6 |
166 |
0.91 |
| chr11_53307549_53307709 | 0.39 |
Hspa4 |
heat shock protein 4 |
7172 |
0.15 |
| chr14_75795151_75795306 | 0.39 |
Slc25a30 |
solute carrier family 25, member 30 |
8191 |
0.15 |
| chr2_5609208_5609400 | 0.39 |
Gm13216 |
predicted gene 13216 |
5733 |
0.28 |
| chr11_49267629_49267816 | 0.38 |
Olfr1393 |
olfactory receptor 1393 |
10582 |
0.11 |
| chr2_132863556_132863823 | 0.38 |
Crls1 |
cardiolipin synthase 1 |
15951 |
0.12 |
| chr18_32254065_32254257 | 0.38 |
Ercc3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
3108 |
0.27 |
| chr10_24106562_24106716 | 0.37 |
Taar9 |
trace amine-associated receptor 9 |
2895 |
0.13 |
| chr4_32561294_32561627 | 0.37 |
Bach2os |
BTB and CNC homology 2, opposite strand |
10157 |
0.16 |
| chr15_3353815_3354005 | 0.37 |
Ccdc152 |
coiled-coil domain containing 152 |
50384 |
0.15 |
| chr8_33853619_33853844 | 0.37 |
Rbpms |
RNA binding protein gene with multiple splicing |
58 |
0.97 |
| chr13_28507894_28508045 | 0.37 |
Gm6081 |
predicted gene 6081 |
3263 |
0.28 |
| chr19_46975101_46975486 | 0.37 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
5725 |
0.17 |
| chr6_108136848_108136999 | 0.37 |
Rpl36-ps12 |
ribosomal protein L36, pseudogene 12 |
3586 |
0.26 |
| chr19_21012514_21012680 | 0.37 |
Tmc1 |
transmembrane channel-like gene family 1 |
58395 |
0.13 |
| chr4_61949215_61949401 | 0.37 |
Mup-ps19 |
major urinary protein, pseudogene 19 |
10603 |
0.17 |
| chr3_81663585_81663896 | 0.37 |
Gm43346 |
predicted gene 43346 |
57936 |
0.14 |
| chr11_20934751_20935121 | 0.36 |
Gm23681 |
predicted gene, 23681 |
2765 |
0.31 |
| chr19_33099137_33099698 | 0.36 |
Gm29946 |
predicted gene, 29946 |
23230 |
0.18 |
| chr19_30151607_30151827 | 0.36 |
Gldc |
glycine decarboxylase |
6486 |
0.2 |
| chr18_31936613_31936956 | 0.36 |
Lims2 |
LIM and senescent cell antigen like domains 2 |
492 |
0.73 |
| chr10_84803969_84804150 | 0.36 |
Gm24226 |
predicted gene, 24226 |
7124 |
0.23 |
| chr2_152397803_152397979 | 0.36 |
Sox12 |
SRY (sex determining region Y)-box 12 |
155 |
0.89 |
| chr5_9032498_9032725 | 0.36 |
Gm40264 |
predicted gene, 40264 |
2169 |
0.24 |
| chr5_89322164_89322355 | 0.35 |
Gc |
vitamin D binding protein |
113369 |
0.07 |
| chr13_38626107_38626269 | 0.35 |
Gm47400 |
predicted gene, 47400 |
8783 |
0.14 |
| chr17_84997546_84997697 | 0.35 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
3941 |
0.22 |
| chr6_121850527_121851087 | 0.35 |
Mug1 |
murinoglobulin 1 |
9703 |
0.21 |
| chr11_59514377_59514532 | 0.35 |
4933439C10Rik |
RIKEN cDNA 4933439C10 gene |
6813 |
0.1 |
| chr6_129539224_129539435 | 0.35 |
Gm44243 |
predicted gene, 44243 |
892 |
0.35 |
| chr13_47154962_47155118 | 0.35 |
Gm48581 |
predicted gene, 48581 |
32 |
0.97 |
| chr8_94070045_94070216 | 0.35 |
Bbs2 |
Bardet-Biedl syndrome 2 (human) |
83 |
0.95 |
| chr9_77845751_77846338 | 0.35 |
Gm19572 |
predicted gene, 19572 |
5664 |
0.15 |
| chr10_59205891_59206053 | 0.35 |
Gm23223 |
predicted gene, 23223 |
10273 |
0.15 |
| chr11_90269750_90269901 | 0.35 |
Mmd |
monocyte to macrophage differentiation-associated |
20349 |
0.21 |
| chr15_36432237_36432390 | 0.34 |
Gm49224 |
predicted gene, 49224 |
4378 |
0.18 |
| chr17_15341268_15341498 | 0.34 |
Gm7423 |
predicted gene 7423 |
19501 |
0.14 |
| chr7_87222899_87223055 | 0.34 |
Gm6230 |
predicted gene 6230 |
19908 |
0.21 |
| chr14_68458990_68459141 | 0.34 |
Gm31227 |
predicted gene, 31227 |
2566 |
0.36 |
| chr9_4372230_4372403 | 0.34 |
Gm23992 |
predicted gene, 23992 |
981 |
0.54 |
| chr12_85366293_85366504 | 0.34 |
Gm47817 |
predicted gene, 47817 |
2753 |
0.16 |
| chr2_72881753_72881905 | 0.34 |
Sp3 |
trans-acting transcription factor 3 |
59255 |
0.1 |
| chr13_37944290_37944811 | 0.33 |
Rreb1 |
ras responsive element binding protein 1 |
2466 |
0.3 |
| chr5_126566218_126566538 | 0.33 |
Gm24839 |
predicted gene, 24839 |
52397 |
0.14 |
| chr4_144977157_144977352 | 0.33 |
Vps13d |
vacuolar protein sorting 13D |
6707 |
0.21 |
| chr8_119436291_119436442 | 0.33 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
752 |
0.59 |
| chr19_29796308_29796471 | 0.33 |
9930021J03Rik |
RIKEN cDNA 9930021J03 gene |
9118 |
0.17 |
| chr1_82202782_82202972 | 0.33 |
Gm9747 |
predicted gene 9747 |
30235 |
0.18 |
| chr15_55252499_55252656 | 0.33 |
Gm26296 |
predicted gene, 26296 |
10026 |
0.23 |
| chr13_94143352_94143530 | 0.33 |
Gm15907 |
predicted gene 15907 |
3324 |
0.24 |
| chr16_20102189_20102498 | 0.33 |
Klhl24 |
kelch-like 24 |
4751 |
0.22 |
| chr4_149978768_149979375 | 0.33 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
22194 |
0.13 |
| chr8_106882742_106882961 | 0.33 |
Chtf8 |
CTF8, chromosome transmission fidelity factor 8 |
10364 |
0.12 |
| chr17_84987973_84988377 | 0.33 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
5498 |
0.2 |
| chr15_66911263_66911477 | 0.33 |
Ccn4 |
cellular communication network factor 4 |
2059 |
0.29 |
| chr19_36534453_36534632 | 0.33 |
Hectd2 |
HECT domain E3 ubiquitin protein ligase 2 |
20097 |
0.2 |
| chr3_107687032_107687196 | 0.32 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
8849 |
0.16 |
| chr1_74344854_74345034 | 0.32 |
Pnkd |
paroxysmal nonkinesiogenic dyskinesia |
12281 |
0.09 |
| chr8_126754096_126754268 | 0.32 |
Gm45805 |
predicted gene 45805 |
4152 |
0.29 |
| chr4_135741644_135741824 | 0.32 |
Il22ra1 |
interleukin 22 receptor, alpha 1 |
13562 |
0.12 |
| chr18_65921955_65922109 | 0.32 |
Gm50118 |
predicted gene, 50118 |
175 |
0.88 |
| chr17_29282347_29282498 | 0.32 |
BC004004 |
cDNA sequence BC004004 |
5740 |
0.11 |
| chr5_146232349_146232681 | 0.32 |
Cdk8 |
cyclin-dependent kinase 8 |
1130 |
0.35 |
| chr15_75596455_75596620 | 0.32 |
Gpihbp1 |
GPI-anchored HDL-binding protein 1 |
91 |
0.95 |
| chr5_8962552_8962757 | 0.32 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
4102 |
0.15 |
| chr4_11896333_11896507 | 0.32 |
Gm25002 |
predicted gene, 25002 |
52467 |
0.11 |
| chr6_4926913_4927078 | 0.32 |
Gm26814 |
predicted gene, 26814 |
15152 |
0.13 |
| chr6_85262797_85262970 | 0.31 |
Sfxn5 |
sideroflexin 5 |
2050 |
0.29 |
| chr7_46840442_46840768 | 0.31 |
Ldha |
lactate dehydrogenase A |
870 |
0.39 |
| chr13_110421353_110421528 | 0.31 |
Plk2 |
polo like kinase 2 |
23643 |
0.22 |
| chr8_86690399_86690763 | 0.31 |
Gm22305 |
predicted gene, 22305 |
19238 |
0.15 |
| chr7_130523668_130523823 | 0.31 |
Gm4275 |
predicted gene 4275 |
10 |
0.96 |
| chr11_86995694_86995863 | 0.31 |
Gm22387 |
predicted gene, 22387 |
1485 |
0.32 |
| chr4_114958381_114958568 | 0.31 |
Cmpk1 |
cytidine monophosphate (UMP-CMP) kinase 1 |
28598 |
0.11 |
| chr16_22914452_22914638 | 0.31 |
Fetub |
fetuin beta |
3789 |
0.14 |
| chr15_79323545_79323705 | 0.31 |
Pla2g6 |
phospholipase A2, group VI |
59 |
0.95 |
| chr2_64973925_64974085 | 0.31 |
Grb14 |
growth factor receptor bound protein 14 |
1741 |
0.5 |
| chr11_108846353_108846520 | 0.31 |
Cep112it |
centrosomal protein 112, intronic transcript |
26103 |
0.19 |
| chr11_106053420_106053645 | 0.31 |
Gm11646 |
predicted gene 11646 |
1680 |
0.21 |
| chr16_91828588_91828869 | 0.31 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
1191 |
0.47 |
| chr6_42251414_42251583 | 0.31 |
Gstk1 |
glutathione S-transferase kappa 1 |
5475 |
0.12 |
| chr14_66748215_66748366 | 0.31 |
Gm10032 |
predicted gene 10032 |
36190 |
0.15 |
| chr7_126260333_126260533 | 0.31 |
Sbk1 |
SH3-binding kinase 1 |
11571 |
0.11 |
| chr6_122174213_122174364 | 0.30 |
Mug-ps1 |
murinoglobulin, pseudogene 1 |
1727 |
0.34 |
| chr18_65075884_65076295 | 0.30 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
57 |
0.98 |
| chr7_114582552_114582724 | 0.30 |
Cyp2r1 |
cytochrome P450, family 2, subfamily r, polypeptide 1 |
19666 |
0.19 |
| chr9_69314244_69314596 | 0.30 |
Rora |
RAR-related orphan receptor alpha |
24738 |
0.18 |
| chr10_77061077_77061274 | 0.30 |
Col18a1 |
collagen, type XVIII, alpha 1 |
1178 |
0.41 |
| chr14_70699508_70699766 | 0.30 |
Xpo7 |
exportin 7 |
8398 |
0.15 |
| chr8_10899350_10899661 | 0.30 |
4833411C07Rik |
RIKEN cDNA 4833411C07 gene |
417 |
0.64 |
| chr2_103846371_103846545 | 0.30 |
Gm13879 |
predicted gene 13879 |
2802 |
0.12 |
| chr11_4986459_4986642 | 0.30 |
Ap1b1 |
adaptor protein complex AP-1, beta 1 subunit |
274 |
0.88 |
| chr6_37748273_37748424 | 0.29 |
Atp6v0c-ps2 |
ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2 |
57738 |
0.12 |
| chr4_33250016_33250167 | 0.29 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
1581 |
0.35 |
| chr4_150748175_150748630 | 0.29 |
Gm16079 |
predicted gene 16079 |
69610 |
0.08 |
| chr5_93008624_93008788 | 0.29 |
Gm25521 |
predicted gene, 25521 |
4433 |
0.16 |
| chr2_103834210_103834381 | 0.29 |
Gm13877 |
predicted gene 13877 |
3222 |
0.11 |
| chr6_97871999_97872401 | 0.29 |
Gm15531 |
predicted gene 15531 |
18339 |
0.23 |
| chr11_16704048_16704199 | 0.29 |
Gm25698 |
predicted gene, 25698 |
28588 |
0.15 |
| chr2_167727310_167727515 | 0.29 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
36231 |
0.09 |
| chr1_59176886_59177061 | 0.29 |
Mpp4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
13584 |
0.12 |
| chr12_54946386_54946548 | 0.29 |
Baz1a |
bromodomain adjacent to zinc finger domain 1A |
2415 |
0.21 |
| chr5_137975596_137975772 | 0.29 |
Azgp1 |
alpha-2-glycoprotein 1, zinc |
5836 |
0.09 |
| chr19_46132112_46132476 | 0.29 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
397 |
0.76 |
| chr5_107413821_107414114 | 0.29 |
Ephx4 |
epoxide hydrolase 4 |
8182 |
0.1 |
| chr12_26460028_26460238 | 0.29 |
Rsad2 |
radical S-adenosyl methionine domain containing 2 |
3681 |
0.17 |
| chr19_21077803_21077989 | 0.29 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
26606 |
0.21 |
| chr15_10872191_10872614 | 0.29 |
Gm29742 |
predicted gene, 29742 |
45035 |
0.12 |
| chr15_39852629_39852953 | 0.28 |
Dpys |
dihydropyrimidinase |
4676 |
0.25 |
| chr18_64755884_64756035 | 0.28 |
Gm23016 |
predicted gene, 23016 |
27893 |
0.12 |
| chrX_167229521_167229672 | 0.28 |
Tmsb4x |
thymosin, beta 4, X chromosome |
20281 |
0.17 |
| chr17_57231348_57231526 | 0.28 |
C3 |
complement component 3 |
3301 |
0.14 |
| chr2_126584960_126585111 | 0.28 |
Hdc |
histidine decarboxylase |
13226 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.7 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.6 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.4 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.5 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.3 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.5 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.4 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.2 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.4 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.2 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.1 | 0.2 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.3 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.2 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.2 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.2 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0046078 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.2 | GO:0014857 | regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 | 0.1 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:1903376 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.0 | GO:0071351 | cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0072610 | interleukin-12 secretion(GO:0072610) |
| 0.0 | 0.0 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.0 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) establishment of T cell polarity(GO:0001768) |
| 0.0 | 0.2 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.0 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.0 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.0 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.0 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.0 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.0 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.0 | 0.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.0 | GO:0002894 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.0 | 0.0 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.1 | GO:0034030 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.0 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.6 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.2 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.1 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.5 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.7 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |