Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
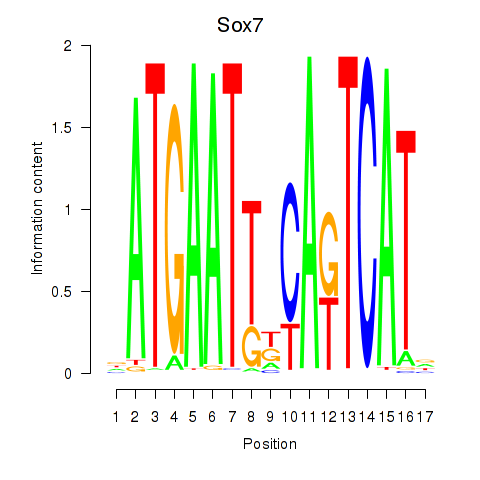
Results for Sox7
Z-value: 1.43
Transcription factors associated with Sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox7
|
ENSMUSG00000063060.5 | SRY (sex determining region Y)-box 7 |
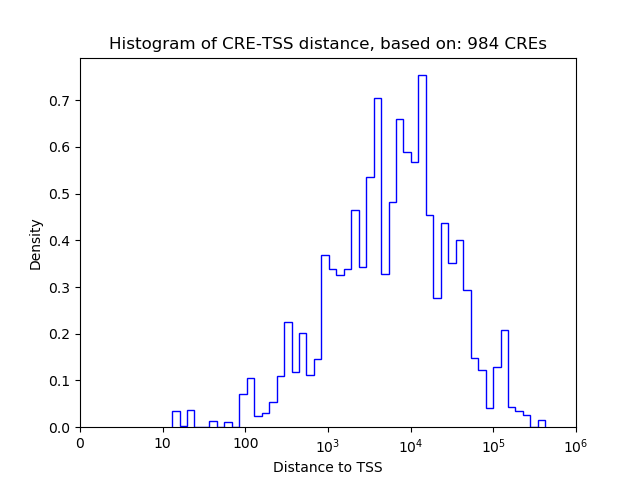
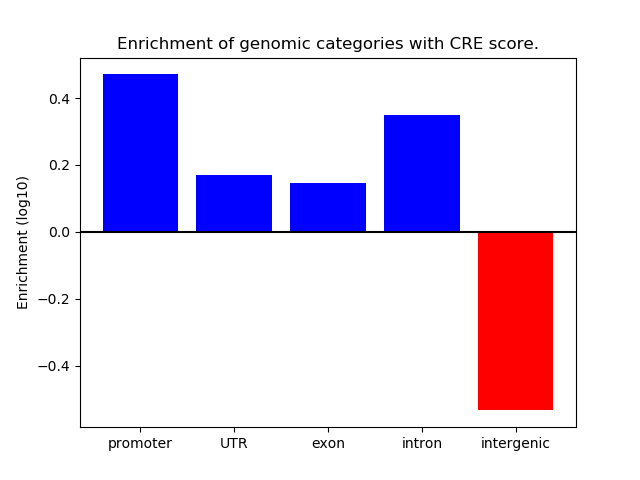
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_63942319_63942611 | Sox7 | 1208 | 0.456001 | 0.85 | 3.2e-02 | Click! |
| chr14_63957868_63958033 | Sox7 | 14277 | 0.157822 | -0.81 | 5.3e-02 | Click! |
| chr14_63941003_63941175 | Sox7 | 2584 | 0.258372 | -0.78 | 6.7e-02 | Click! |
| chr14_63943165_63943771 | Sox7 | 205 | 0.936046 | -0.74 | 9.0e-02 | Click! |
| chr14_63941556_63941788 | Sox7 | 2001 | 0.305936 | 0.71 | 1.1e-01 | Click! |
Activity of the Sox7 motif across conditions
Conditions sorted by the z-value of the Sox7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
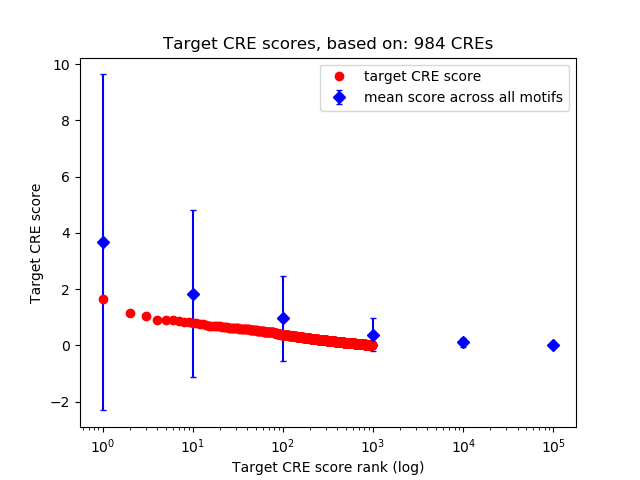
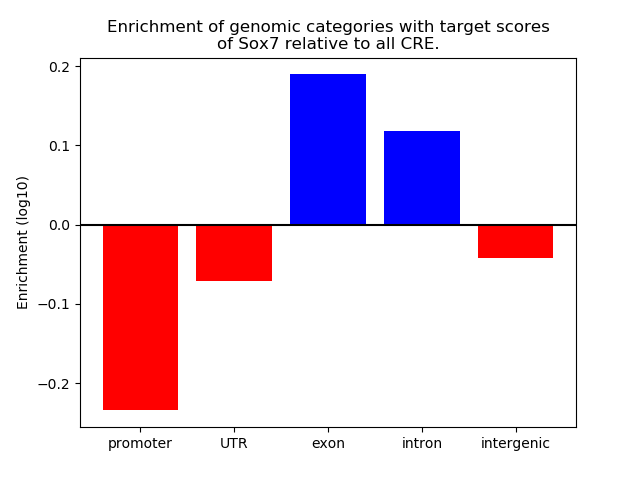
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_127899545_127899941 | 1.63 |
Rab3gap1 |
RAB3 GTPase activating protein subunit 1 |
1170 |
0.47 |
| chr7_114735164_114735333 | 1.15 |
Insc |
INSC spindle orientation adaptor protein |
8446 |
0.16 |
| chr4_22498530_22498747 | 1.03 |
Gm30731 |
predicted gene, 30731 |
8090 |
0.16 |
| chr9_45204142_45204293 | 0.92 |
Tmprss4 |
transmembrane protease, serine 4 |
125 |
0.93 |
| chr4_44920931_44921086 | 0.91 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
2110 |
0.25 |
| chr1_67211421_67211668 | 0.90 |
Gm15668 |
predicted gene 15668 |
37656 |
0.16 |
| chr5_32243149_32243309 | 0.85 |
Plb1 |
phospholipase B1 |
4122 |
0.22 |
| chr14_21102558_21102733 | 0.83 |
Adk |
adenosine kinase |
26493 |
0.19 |
| chr10_87884302_87884648 | 0.82 |
Igf1os |
insulin-like growth factor 1, opposite strand |
21094 |
0.18 |
| chr2_70813245_70813396 | 0.81 |
Tlk1 |
tousled-like kinase 1 |
11908 |
0.21 |
| chr8_104789386_104789560 | 0.79 |
Gm45782 |
predicted gene 45782 |
3645 |
0.12 |
| chr1_130734318_130734676 | 0.77 |
AA986860 |
expressed sequence AA986860 |
2387 |
0.14 |
| chr1_67160278_67160544 | 0.75 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
37385 |
0.17 |
| chr8_93183400_93183567 | 0.72 |
Gm45909 |
predicted gene 45909 |
7875 |
0.13 |
| chr7_113961934_113962117 | 0.70 |
Gm45615 |
predicted gene 45615 |
124873 |
0.05 |
| chr2_134947281_134947437 | 0.69 |
Gm14036 |
predicted gene 14036 |
143410 |
0.04 |
| chr4_63360320_63360510 | 0.68 |
Orm2 |
orosomucoid 2 |
2034 |
0.19 |
| chr13_96592864_96593019 | 0.68 |
Gm48139 |
predicted gene, 48139 |
12736 |
0.12 |
| chr4_47366562_47366857 | 0.67 |
Tgfbr1 |
transforming growth factor, beta receptor I |
13099 |
0.22 |
| chr6_27782346_27782517 | 0.67 |
Gm26310 |
predicted gene, 26310 |
114344 |
0.07 |
| chr10_87913707_87914126 | 0.67 |
Igf1os |
insulin-like growth factor 1, opposite strand |
50535 |
0.11 |
| chr15_94674453_94674604 | 0.66 |
Gm25546 |
predicted gene, 25546 |
6858 |
0.25 |
| chr12_104342354_104343298 | 0.66 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
4340 |
0.13 |
| chr2_118307958_118308132 | 0.65 |
1700054M17Rik |
RIKEN cDNA 1700054M17 gene |
3131 |
0.19 |
| chr15_3482890_3483092 | 0.63 |
Ghr |
growth hormone receptor |
11347 |
0.28 |
| chr4_107313475_107313626 | 0.63 |
Yipf1 |
Yip1 domain family, member 1 |
813 |
0.49 |
| chr9_66699874_66700052 | 0.63 |
Car12 |
carbonic anhydrase 12 |
13723 |
0.2 |
| chr18_33436230_33436484 | 0.63 |
Nrep |
neuronal regeneration related protein |
27078 |
0.18 |
| chr6_23387093_23387244 | 0.62 |
Cadps2 |
Ca2+-dependent activator protein for secretion 2 |
31249 |
0.21 |
| chr8_22860131_22860751 | 0.62 |
Kat6a |
K(lysine) acetyltransferase 6A |
892 |
0.52 |
| chr2_155062362_155062698 | 0.61 |
Gm45609 |
predicted gene 45609 |
11651 |
0.13 |
| chr8_25750286_25750467 | 0.60 |
Ddhd2 |
DDHD domain containing 2 |
526 |
0.61 |
| chr8_42311444_42311600 | 0.60 |
Gm6180 |
predicted pseudogene 6180 |
64611 |
0.15 |
| chr15_54578033_54578470 | 0.59 |
Mal2 |
mal, T cell differentiation protein 2 |
7059 |
0.27 |
| chr9_45958350_45958531 | 0.59 |
Sidt2 |
SID1 transmembrane family, member 2 |
3182 |
0.13 |
| chr6_13676526_13676747 | 0.59 |
Bmt2 |
base methyltransferase of 25S rRNA 2 |
1302 |
0.55 |
| chr19_39346810_39347020 | 0.59 |
Cyp2c72-ps |
cytochrome P450, family 2, subfamily c, polypeptide 72, pseudogene |
452 |
0.87 |
| chr11_106468981_106469217 | 0.59 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
18697 |
0.14 |
| chr8_26350253_26350419 | 0.58 |
Gm31784 |
predicted gene, 31784 |
38002 |
0.12 |
| chr16_43535731_43535882 | 0.57 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
25498 |
0.2 |
| chr5_20858526_20858689 | 0.57 |
Phtf2 |
putative homeodomain transcription factor 2 |
3416 |
0.23 |
| chr3_105737986_105738138 | 0.56 |
Gm43328 |
predicted gene 43328 |
4185 |
0.13 |
| chr3_62340540_62340713 | 0.56 |
9330121J05Rik |
RIKEN cDNA 9330121J05 gene |
94 |
0.7 |
| chr11_29130922_29131328 | 0.56 |
Pnpt1 |
polyribonucleotide nucleotidyltransferase 1 |
377 |
0.88 |
| chr17_84742495_84742888 | 0.56 |
Lrpprc |
leucine-rich PPR-motif containing |
8571 |
0.17 |
| chr6_52009095_52009246 | 0.55 |
Skap2 |
src family associated phosphoprotein 2 |
3345 |
0.2 |
| chr11_16867245_16867846 | 0.55 |
Egfr |
epidermal growth factor receptor |
10605 |
0.2 |
| chr4_117131790_117132241 | 0.55 |
Plk3 |
polo like kinase 3 |
1784 |
0.13 |
| chr5_130015627_130015778 | 0.55 |
Asl |
argininosuccinate lyase |
980 |
0.41 |
| chr13_91748092_91748287 | 0.54 |
Gm27656 |
predicted gene, 27656 |
3917 |
0.17 |
| chr5_91039831_91039982 | 0.54 |
Epgn |
epithelial mitogen |
12442 |
0.17 |
| chr6_117360291_117360442 | 0.53 |
Gm4640 |
predicted gene 4640 |
63251 |
0.12 |
| chr8_127515465_127515616 | 0.53 |
Gm6921 |
predicted pseudogene 6921 |
45895 |
0.17 |
| chr7_63899068_63899509 | 0.51 |
Gm27252 |
predicted gene 27252 |
1314 |
0.37 |
| chr4_40145230_40145427 | 0.51 |
Aco1 |
aconitase 1 |
2247 |
0.3 |
| chr6_149150480_149150842 | 0.51 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
9015 |
0.13 |
| chr15_82433343_82433494 | 0.50 |
Cyp2d9 |
cytochrome P450, family 2, subfamily d, polypeptide 9 |
19 |
0.57 |
| chr9_119309703_119309944 | 0.50 |
Gm22729 |
predicted gene, 22729 |
3066 |
0.17 |
| chr2_146311135_146311471 | 0.50 |
Gm14117 |
predicted gene 14117 |
5756 |
0.24 |
| chr1_63122681_63122832 | 0.49 |
Gm20342 |
predicted gene, 20342 |
5638 |
0.1 |
| chr8_95751194_95751345 | 0.49 |
Mir7073 |
microRNA 7073 |
1945 |
0.17 |
| chr3_82036127_82036278 | 0.49 |
Gucy1b1 |
guanylate cyclase 1, soluble, beta 1 |
880 |
0.6 |
| chr1_119648022_119648226 | 0.48 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
493 |
0.78 |
| chr4_102583510_102583771 | 0.48 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
3918 |
0.35 |
| chr4_71811423_71811574 | 0.48 |
Gm11232 |
predicted gene 11232 |
53533 |
0.15 |
| chr2_122158672_122158840 | 0.48 |
Trim69 |
tripartite motif-containing 69 |
1944 |
0.21 |
| chr4_12481585_12481744 | 0.48 |
Gm37985 |
predicted gene, 37985 |
115724 |
0.06 |
| chr4_9665988_9666139 | 0.48 |
Asph |
aspartate-beta-hydroxylase |
3023 |
0.24 |
| chr9_32870948_32871133 | 0.47 |
Gm37167 |
predicted gene, 37167 |
31118 |
0.16 |
| chr9_62357318_62357469 | 0.47 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
9545 |
0.21 |
| chr2_31511401_31511572 | 0.47 |
Ass1 |
argininosuccinate synthetase 1 |
7004 |
0.18 |
| chr16_35491110_35491284 | 0.47 |
Pdia5 |
protein disulfide isomerase associated 5 |
324 |
0.87 |
| chr4_15066667_15066833 | 0.47 |
Gm11844 |
predicted gene 11844 |
44168 |
0.16 |
| chr16_42998828_42999221 | 0.46 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43393 |
0.16 |
| chr5_122494527_122494678 | 0.46 |
Gm43360 |
predicted gene 43360 |
306 |
0.77 |
| chr6_31551331_31551519 | 0.46 |
Podxl |
podocalyxin-like |
12556 |
0.18 |
| chr14_59205202_59205353 | 0.46 |
Rcbtb1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
336 |
0.88 |
| chr13_81212164_81212315 | 0.46 |
Gm48566 |
predicted gene, 48566 |
28545 |
0.2 |
| chr7_110816193_110816374 | 0.45 |
Rnf141 |
ring finger protein 141 |
5000 |
0.16 |
| chr15_5106677_5106828 | 0.45 |
Card6 |
caspase recruitment domain family, member 6 |
1732 |
0.22 |
| chr4_108095142_108095451 | 0.44 |
Podn |
podocan |
1149 |
0.41 |
| chr16_76371516_76371667 | 0.44 |
Nrip1 |
nuclear receptor interacting protein 1 |
1446 |
0.43 |
| chr2_34775525_34776380 | 0.43 |
Hspa5 |
heat shock protein 5 |
1105 |
0.4 |
| chr1_100410526_100410677 | 0.43 |
Gm29667 |
predicted gene 29667 |
127810 |
0.05 |
| chr17_30042343_30042527 | 0.42 |
Zfand3 |
zinc finger, AN1-type domain 3 |
17947 |
0.15 |
| chr4_41331357_41331705 | 0.41 |
Gm26084 |
predicted gene, 26084 |
14420 |
0.1 |
| chr1_65170171_65170540 | 0.41 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
5209 |
0.15 |
| chr5_122494845_122494996 | 0.41 |
Gm43360 |
predicted gene 43360 |
12 |
0.94 |
| chr10_63038751_63038902 | 0.39 |
Pbld2 |
phenazine biosynthesis-like protein domain containing 2 |
14314 |
0.09 |
| chr16_46853625_46853776 | 0.39 |
Gm6912 |
predicted gene 6912 |
220498 |
0.02 |
| chr2_11427432_11427583 | 0.39 |
Gm13296 |
predicted gene 13296 |
7283 |
0.12 |
| chr3_133740354_133740509 | 0.39 |
Gm6135 |
prediticted gene 6135 |
51073 |
0.13 |
| chr11_32681512_32681693 | 0.39 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
38698 |
0.16 |
| chr11_106429328_106429479 | 0.39 |
Icam2 |
intercellular adhesion molecule 2 |
41328 |
0.1 |
| chr6_115653735_115653886 | 0.39 |
Gm14335 |
predicted gene 14335 |
14751 |
0.11 |
| chr6_67268636_67268787 | 0.38 |
Serbp1 |
serpine1 mRNA binding protein 1 |
1381 |
0.34 |
| chr4_48272957_48273115 | 0.38 |
Erp44 |
endoplasmic reticulum protein 44 |
6416 |
0.2 |
| chr19_43808546_43808716 | 0.38 |
Abcc2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
1891 |
0.28 |
| chr3_52006994_52007318 | 0.37 |
Gm37465 |
predicted gene, 37465 |
3131 |
0.17 |
| chr18_20554666_20554835 | 0.37 |
Dsg2 |
desmoglein 2 |
3324 |
0.21 |
| chr1_164435834_164435997 | 0.37 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
7804 |
0.15 |
| chr1_127787315_127787490 | 0.37 |
Ccnt2 |
cyclin T2 |
9169 |
0.16 |
| chr3_24123037_24123188 | 0.37 |
Naaladl2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
150927 |
0.05 |
| chr3_36480639_36480833 | 0.37 |
1810062G17Rik |
RIKEN cDNA 1810062G17 gene |
4799 |
0.13 |
| chr8_105090981_105091315 | 0.36 |
Ces3b |
carboxylesterase 3B |
2529 |
0.16 |
| chr3_135442828_135442979 | 0.36 |
Ube2d3 |
ubiquitin-conjugating enzyme E2D 3 |
3692 |
0.12 |
| chr9_76727753_76728144 | 0.36 |
Gm22938 |
predicted gene, 22938 |
11201 |
0.17 |
| chr3_84234361_84234584 | 0.36 |
Trim2 |
tripartite motif-containing 2 |
13583 |
0.23 |
| chr6_17478662_17478867 | 0.35 |
Met |
met proto-oncogene |
12477 |
0.21 |
| chr2_148031539_148031764 | 0.35 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
6619 |
0.18 |
| chr9_92257106_92257271 | 0.35 |
Plscr1 |
phospholipid scramblase 1 |
6846 |
0.15 |
| chr13_9495114_9495298 | 0.35 |
Gm48871 |
predicted gene, 48871 |
49306 |
0.11 |
| chr13_4061646_4061992 | 0.35 |
Akr1c14 |
aldo-keto reductase family 1, member C14 |
2228 |
0.22 |
| chr1_51856674_51857057 | 0.35 |
Myo1b |
myosin IB |
5759 |
0.17 |
| chr6_29852245_29852396 | 0.35 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
1440 |
0.41 |
| chr17_78379527_78379767 | 0.35 |
Fez2 |
fasciculation and elongation protein zeta 2 (zygin II) |
21783 |
0.15 |
| chr1_67127092_67127716 | 0.34 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
4378 |
0.28 |
| chr5_146293255_146293729 | 0.34 |
Cdk8 |
cyclin-dependent kinase 8 |
1418 |
0.38 |
| chr10_5408928_5409079 | 0.34 |
Syne1 |
spectrin repeat containing, nuclear envelope 1 |
40517 |
0.18 |
| chr12_8694008_8694159 | 0.34 |
Pum2 |
pumilio RNA-binding family member 2 |
4013 |
0.27 |
| chr11_115219587_115219763 | 0.34 |
Nat9 |
N-acetyltransferase 9 (GCN5-related, putative) |
31816 |
0.08 |
| chr16_84837970_84838147 | 0.34 |
Atp5j |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
2433 |
0.2 |
| chr8_125493583_125493749 | 0.34 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
956 |
0.69 |
| chr11_60248805_60248956 | 0.34 |
Tom1l2 |
target of myb1-like 2 (chicken) |
3211 |
0.16 |
| chr19_40141682_40141833 | 0.33 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
45529 |
0.11 |
| chr7_115680029_115680180 | 0.33 |
Sox6 |
SRY (sex determining region Y)-box 6 |
17739 |
0.29 |
| chr3_130339266_130339443 | 0.33 |
Gm22682 |
predicted gene, 22682 |
133889 |
0.04 |
| chr14_11158870_11159044 | 0.33 |
Slc25a5-ps |
Slc25a5 retrotransposed pseudogene |
2572 |
0.28 |
| chr13_4028561_4028716 | 0.33 |
Gm47853 |
predicted gene, 47853 |
5469 |
0.14 |
| chr12_79435773_79435986 | 0.32 |
Rad51b |
RAD51 paralog B |
108526 |
0.06 |
| chr4_134917124_134917439 | 0.32 |
Tmem50a |
transmembrane protein 50A |
2257 |
0.24 |
| chr12_80124026_80124199 | 0.32 |
2310015A10Rik |
RIKEN cDNA 2310015A10 gene |
2973 |
0.17 |
| chr15_3453354_3453796 | 0.32 |
Ghr |
growth hormone receptor |
18069 |
0.26 |
| chr10_17946556_17946707 | 0.32 |
Heca |
hdc homolog, cell cycle regulator |
1436 |
0.47 |
| chr16_11176623_11176798 | 0.32 |
Zc3h7a |
zinc finger CCCH type containing 7 A |
317 |
0.74 |
| chr15_97025258_97025453 | 0.32 |
Slc38a4 |
solute carrier family 38, member 4 |
5033 |
0.31 |
| chr6_35338820_35339318 | 0.32 |
1700065J11Rik |
RIKEN cDNA 1700065J11 gene |
8223 |
0.18 |
| chr2_43592187_43592360 | 0.32 |
Kynu |
kynureninase |
7342 |
0.3 |
| chr9_46481072_46481223 | 0.31 |
Gm47144 |
predicted gene, 47144 |
23926 |
0.16 |
| chr1_71595444_71595633 | 0.31 |
Fn1 |
fibronectin 1 |
4467 |
0.23 |
| chr11_18900021_18900190 | 0.31 |
2900018N21Rik |
RIKEN cDNA 2900018N21 gene |
10265 |
0.16 |
| chr4_19970512_19970663 | 0.31 |
4930480G23Rik |
RIKEN cDNA 4930480G23 gene |
27649 |
0.17 |
| chr5_136258958_136259125 | 0.31 |
Sh2b2 |
SH2B adaptor protein 2 |
12485 |
0.13 |
| chr5_17998832_17999006 | 0.31 |
Gnat3 |
guanine nucleotide binding protein, alpha transducing 3 |
36370 |
0.22 |
| chr19_44394517_44394772 | 0.31 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12046 |
0.14 |
| chr4_108849580_108849741 | 0.31 |
Kti12 |
KTI12 homolog, chromatin associated |
1875 |
0.25 |
| chr7_66075917_66076093 | 0.30 |
Gm45081 |
predicted gene 45081 |
3468 |
0.14 |
| chr19_59979470_59979621 | 0.30 |
Gm4219 |
predicted gene 4219 |
35823 |
0.15 |
| chr5_23578433_23578591 | 0.30 |
Srpk2 |
serine/arginine-rich protein specific kinase 2 |
29142 |
0.15 |
| chr10_69218422_69218684 | 0.30 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
832 |
0.61 |
| chr3_95324375_95324754 | 0.30 |
Cers2 |
ceramide synthase 2 |
3624 |
0.1 |
| chr8_47712885_47713036 | 0.30 |
E030037K01Rik |
RIKEN cDNA E030037K01 gene |
306 |
0.75 |
| chr13_96668366_96668901 | 0.30 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
1021 |
0.47 |
| chr7_127277970_127278299 | 0.30 |
Sephs2 |
selenophosphate synthetase 2 |
4079 |
0.08 |
| chr6_93770467_93770664 | 0.30 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
21974 |
0.21 |
| chr2_59169467_59169618 | 0.30 |
Pkp4 |
plakophilin 4 |
7615 |
0.2 |
| chr3_60582048_60582207 | 0.29 |
Mbnl1 |
muscleblind like splicing factor 1 |
13514 |
0.2 |
| chr13_119378503_119378661 | 0.29 |
Nnt |
nicotinamide nucleotide transhydrogenase |
30370 |
0.15 |
| chr2_31515291_31515653 | 0.28 |
Ass1 |
argininosuccinate synthetase 1 |
3018 |
0.23 |
| chr9_74790782_74790933 | 0.28 |
Gm22315 |
predicted gene, 22315 |
8787 |
0.19 |
| chr1_57386674_57386899 | 0.28 |
Tyw5 |
tRNA-yW synthesizing protein 5 |
3985 |
0.16 |
| chr4_92376164_92376339 | 0.28 |
Gm12638 |
predicted gene 12638 |
2903 |
0.4 |
| chr10_67191188_67191347 | 0.28 |
Jmjd1c |
jumonji domain containing 1C |
5514 |
0.23 |
| chr9_65748388_65748657 | 0.28 |
Zfp609 |
zinc finger protein 609 |
47378 |
0.11 |
| chr11_16767286_16767565 | 0.28 |
Egfr |
epidermal growth factor receptor |
15195 |
0.19 |
| chr1_168242978_168243129 | 0.28 |
Pbx1 |
pre B cell leukemia homeobox 1 |
78391 |
0.1 |
| chr12_57444721_57445109 | 0.28 |
Gm16246 |
predicted gene 16246 |
5205 |
0.25 |
| chr3_97616365_97616788 | 0.28 |
Chd1l |
chromodomain helicase DNA binding protein 1-like |
6373 |
0.15 |
| chr11_117359987_117360146 | 0.28 |
Septin9 |
septin 9 |
706 |
0.68 |
| chr9_120776284_120776475 | 0.28 |
Gm47066 |
predicted gene, 47066 |
15607 |
0.11 |
| chr15_6891673_6891824 | 0.28 |
Osmr |
oncostatin M receptor |
16779 |
0.26 |
| chr16_22241727_22242034 | 0.28 |
Gm36796 |
predicted gene, 36796 |
2288 |
0.19 |
| chr7_99201987_99202161 | 0.28 |
Gm45012 |
predicted gene 45012 |
292 |
0.86 |
| chr17_64767271_64767462 | 0.27 |
Dreh |
down-regulated in hepatocellular carcinoma |
255 |
0.93 |
| chr1_135210155_135210339 | 0.27 |
Gm4204 |
predicted gene 4204 |
21748 |
0.1 |
| chr3_133833846_133834006 | 0.27 |
Gm30484 |
predicted gene, 30484 |
15408 |
0.23 |
| chr17_32348126_32348277 | 0.27 |
Akap8l |
A kinase (PRKA) anchor protein 8-like |
2305 |
0.19 |
| chr10_87718160_87718329 | 0.27 |
Gm48195 |
predicted gene, 48195 |
29666 |
0.21 |
| chr11_16862337_16862489 | 0.26 |
Egfr |
epidermal growth factor receptor |
15737 |
0.19 |
| chr7_89401915_89402333 | 0.26 |
Tmem135 |
transmembrane protein 135 |
2098 |
0.22 |
| chr19_57272214_57272473 | 0.26 |
4930449E18Rik |
RIKEN cDNA 4930449E18 gene |
1617 |
0.39 |
| chr18_12635155_12635343 | 0.26 |
Ttc39c |
tetratricopeptide repeat domain 39C |
8095 |
0.15 |
| chr18_64628060_64628409 | 0.26 |
Gm6978 |
predicted gene 6978 |
15135 |
0.14 |
| chr6_119390969_119391120 | 0.26 |
Adipor2 |
adiponectin receptor 2 |
796 |
0.67 |
| chr15_31230889_31231047 | 0.26 |
Dap |
death-associated protein |
5772 |
0.19 |
| chr13_52188692_52188881 | 0.25 |
Gm48199 |
predicted gene, 48199 |
8375 |
0.27 |
| chr7_35142488_35142656 | 0.25 |
Gm35665 |
predicted gene, 35665 |
1123 |
0.34 |
| chr17_13687760_13687927 | 0.25 |
Gm16046 |
predicted gene 16046 |
4327 |
0.17 |
| chrX_136321797_136321962 | 0.25 |
Nsa2-ps2 |
NSA2 ribosome biogenesis homolog, pseudogene 2 |
6420 |
0.09 |
| chr2_35103269_35103589 | 0.25 |
AI182371 |
expressed sequence AI182371 |
1886 |
0.29 |
| chr14_68297607_68297758 | 0.25 |
Gm47212 |
predicted gene, 47212 |
79429 |
0.1 |
| chr11_60837742_60838309 | 0.25 |
Dhrs7b |
dehydrogenase/reductase (SDR family) member 7B |
7346 |
0.1 |
| chr3_105758672_105758857 | 0.25 |
Rap1a |
RAS-related protein 1a |
8436 |
0.11 |
| chr1_60616128_60616279 | 0.25 |
Gm23762 |
predicted gene, 23762 |
2400 |
0.22 |
| chr1_166153672_166153856 | 0.25 |
Gpa33 |
glycoprotein A33 (transmembrane) |
23297 |
0.15 |
| chr12_79599250_79599471 | 0.25 |
Rad51b |
RAD51 paralog B |
272007 |
0.01 |
| chr6_56795662_56795822 | 0.25 |
Kbtbd2 |
kelch repeat and BTB (POZ) domain containing 2 |
1005 |
0.43 |
| chr1_84300039_84300190 | 0.25 |
Pid1 |
phosphotyrosine interaction domain containing 1 |
15469 |
0.24 |
| chr13_4205947_4206185 | 0.25 |
Akr1c13 |
aldo-keto reductase family 1, member C13 |
12208 |
0.13 |
| chr11_86739271_86739543 | 0.25 |
Cltc |
clathrin, heavy polypeptide (Hc) |
12924 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 0.5 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.2 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.0 | GO:0035935 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:0006188 | IMP biosynthetic process(GO:0006188) 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |