Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
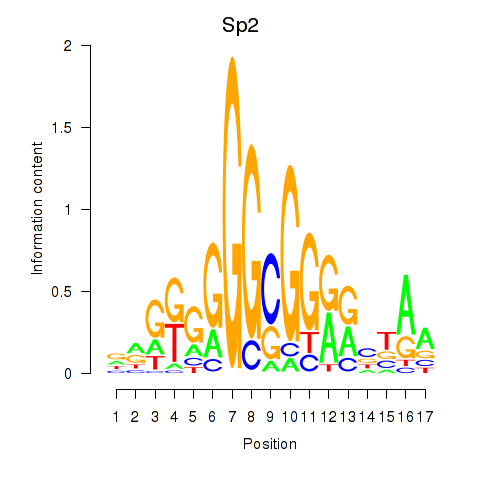
Results for Sp2
Z-value: 1.04
Transcription factors associated with Sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp2
|
ENSMUSG00000018678.6 | Sp2 transcription factor |
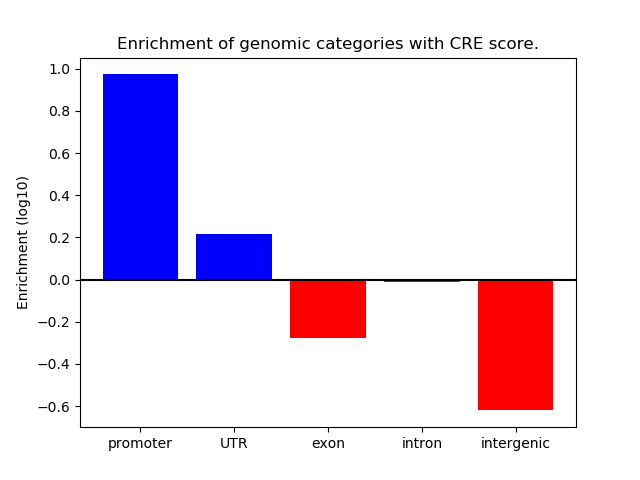
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_96967452_96967847 | Sp2 | 1016 | 0.330273 | -0.86 | 2.7e-02 | Click! |
| chr11_96976103_96976321 | Sp2 | 1499 | 0.218452 | 0.82 | 4.8e-02 | Click! |
| chr11_96963893_96964044 | Sp2 | 4697 | 0.100484 | -0.69 | 1.3e-01 | Click! |
| chr11_96977958_96978364 | Sp2 | 450 | 0.662278 | 0.63 | 1.8e-01 | Click! |
| chr11_96982500_96982661 | Sp2 | 379 | 0.722182 | -0.62 | 1.9e-01 | Click! |
Activity of the Sp2 motif across conditions
Conditions sorted by the z-value of the Sp2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
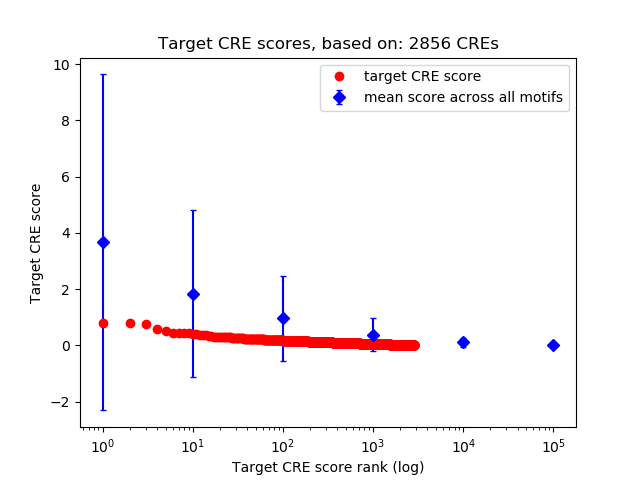
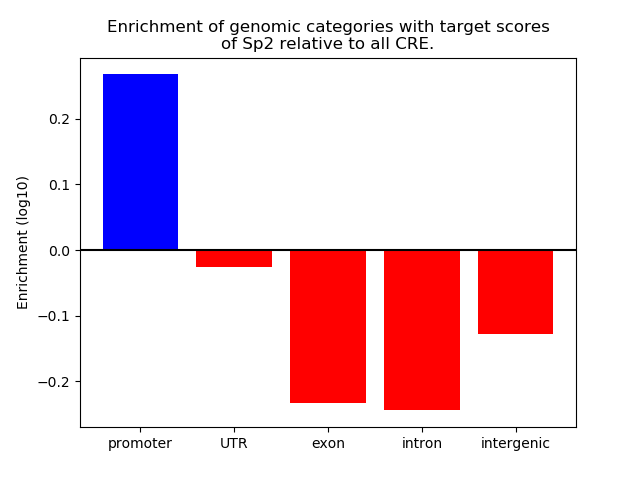
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_98767825_98767976 | 0.78 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2526 |
0.15 |
| chr4_136090324_136090475 | 0.78 |
Gm13009 |
predicted gene 13009 |
15427 |
0.12 |
| chr11_98767109_98767369 | 0.77 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3187 |
0.13 |
| chr3_121982194_121982345 | 0.57 |
Arhgap29 |
Rho GTPase activating protein 29 |
719 |
0.68 |
| chr10_75835778_75835936 | 0.50 |
Gstt2 |
glutathione S-transferase, theta 2 |
976 |
0.36 |
| chr8_119417830_119417981 | 0.45 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
16219 |
0.14 |
| chr6_145865476_145865643 | 0.44 |
Bhlhe41 |
basic helix-loop-helix family, member e41 |
1 |
0.97 |
| chr6_145855716_145856207 | 0.44 |
Gm43909 |
predicted gene, 43909 |
7336 |
0.17 |
| chr18_21300208_21300569 | 0.43 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
250 |
0.91 |
| chr16_45945391_45945574 | 0.42 |
Gm15638 |
predicted gene 15638 |
124 |
0.96 |
| chr7_127477990_127478144 | 0.40 |
Fbrs |
fibrosin |
1132 |
0.21 |
| chr8_122497127_122497278 | 0.38 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
1002 |
0.33 |
| chr2_25983198_25983706 | 0.36 |
Camsap1 |
calmodulin regulated spectrin-associated protein 1 |
170 |
0.93 |
| chr18_64265864_64266047 | 0.35 |
St8sia3 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
31 |
0.97 |
| chr8_108710919_108711070 | 0.34 |
Zfhx3 |
zinc finger homeobox 3 |
3650 |
0.29 |
| chr9_74875936_74876087 | 0.33 |
Onecut1 |
one cut domain, family member 1 |
9527 |
0.15 |
| chr1_74807002_74807292 | 0.31 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
13784 |
0.11 |
| chr11_95032963_95033336 | 0.31 |
Gm11513 |
predicted gene 11513 |
390 |
0.76 |
| chr5_107260769_107260925 | 0.31 |
Gm42900 |
predicted gene 42900 |
4343 |
0.17 |
| chr17_86165279_86165510 | 0.30 |
Prkce |
protein kinase C, epsilon |
2391 |
0.3 |
| chr11_98953496_98953709 | 0.30 |
Rara |
retinoic acid receptor, alpha |
6810 |
0.11 |
| chr18_34864196_34864456 | 0.30 |
Egr1 |
early growth response 1 |
3119 |
0.18 |
| chr8_72492767_72492930 | 0.29 |
Gm17435 |
predicted gene, 17435 |
86 |
0.69 |
| chr5_121836951_121837292 | 0.29 |
Sh2b3 |
SH2B adaptor protein 3 |
262 |
0.83 |
| chr11_117875729_117875880 | 0.28 |
Tha1 |
threonine aldolase 1 |
2323 |
0.16 |
| chr6_137142914_137143065 | 0.28 |
Rerg |
RAS-like, estrogen-regulated, growth-inhibitor |
2926 |
0.27 |
| chr12_105008860_105009027 | 0.28 |
Syne3 |
spectrin repeat containing, nuclear envelope family member 3 |
866 |
0.44 |
| chr19_57877445_57877626 | 0.27 |
Mir5623 |
microRNA 5623 |
173632 |
0.03 |
| chr13_109485029_109485208 | 0.26 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
42935 |
0.21 |
| chr12_86836691_86837004 | 0.26 |
Gm10095 |
predicted gene 10095 |
9620 |
0.19 |
| chr4_149393707_149393902 | 0.26 |
Ube4b |
ubiquitination factor E4B |
5574 |
0.15 |
| chr16_31452530_31452681 | 0.25 |
Bdh1 |
3-hydroxybutyrate dehydrogenase, type 1 |
5511 |
0.15 |
| chr3_41580995_41581174 | 0.25 |
Jade1 |
jade family PHD finger 1 |
228 |
0.92 |
| chr6_121064341_121064666 | 0.25 |
Gm4651 |
predicted gene 4651 |
39 |
0.97 |
| chr18_34511486_34511736 | 0.25 |
n-R5s24 |
nuclear encoded rRNA 5S 24 |
1874 |
0.27 |
| chr9_24774302_24774460 | 0.24 |
Tbx20 |
T-box 20 |
78 |
0.97 |
| chr19_7196833_7197251 | 0.24 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
3422 |
0.16 |
| chr7_25249677_25249858 | 0.24 |
Erf |
Ets2 repressor factor |
963 |
0.32 |
| chr1_9967919_9968070 | 0.24 |
Tcf24 |
transcription factor 24 |
62 |
0.9 |
| chr15_102029131_102029282 | 0.23 |
Krt18 |
keratin 18 |
1026 |
0.4 |
| chr3_142749520_142749995 | 0.23 |
Gm15540 |
predicted gene 15540 |
5076 |
0.14 |
| chr11_62076764_62076946 | 0.23 |
Specc1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
140 |
0.96 |
| chr1_34389167_34389350 | 0.23 |
Gm5266 |
predicted gene 5266 |
21887 |
0.09 |
| chr8_45631075_45631340 | 0.23 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
3006 |
0.26 |
| chr14_45056454_45056605 | 0.23 |
Gm21010 |
predicted gene, 21010 |
34086 |
0.09 |
| chr11_89937352_89937519 | 0.23 |
Pctp |
phosphatidylcholine transfer protein |
65459 |
0.12 |
| chr2_29973635_29973786 | 0.23 |
Sptan1 |
spectrin alpha, non-erythrocytic 1 |
4670 |
0.13 |
| chr9_107675603_107675819 | 0.23 |
Gnat1 |
guanine nucleotide binding protein, alpha transducing 1 |
730 |
0.43 |
| chr7_45053316_45053568 | 0.23 |
Prr12 |
proline rich 12 |
561 |
0.42 |
| chr9_62351261_62351449 | 0.23 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
6556 |
0.21 |
| chr1_175692768_175692950 | 0.22 |
Chml |
choroideremia-like |
42 |
0.54 |
| chr9_42507207_42507377 | 0.22 |
Tbcel |
tubulin folding cofactor E-like |
517 |
0.81 |
| chr8_84723608_84723805 | 0.22 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
699 |
0.52 |
| chr2_128817774_128817954 | 0.22 |
Tmem87b |
transmembrane protein 87B |
254 |
0.9 |
| chr5_88796583_88796899 | 0.22 |
Gm42912 |
predicted gene 42912 |
2845 |
0.21 |
| chr11_109611461_109611655 | 0.22 |
Wipi1 |
WD repeat domain, phosphoinositide interacting 1 |
126 |
0.95 |
| chr9_121380419_121380796 | 0.22 |
Trak1 |
trafficking protein, kinesin binding 1 |
2187 |
0.29 |
| chr8_70539199_70539597 | 0.22 |
Ell |
elongation factor RNA polymerase II |
59 |
0.94 |
| chr15_101266552_101266884 | 0.21 |
Nr4a1 |
nuclear receptor subfamily 4, group A, member 1 |
128 |
0.93 |
| chr9_25455109_25455478 | 0.21 |
Gm8049 |
predicted gene 8049 |
4831 |
0.23 |
| chrX_100447058_100447240 | 0.21 |
Awat2 |
acyl-CoA wax alcohol acyltransferase 2 |
4432 |
0.18 |
| chr6_113306564_113306942 | 0.21 |
Gm43935 |
predicted gene, 43935 |
79 |
0.8 |
| chr11_76911521_76911684 | 0.21 |
Tmigd1 |
transmembrane and immunoglobulin domain containing 1 |
7040 |
0.17 |
| chr15_97029364_97029596 | 0.21 |
Slc38a4 |
solute carrier family 38, member 4 |
1731 |
0.48 |
| chr7_26833022_26833239 | 0.20 |
Cyp2a5 |
cytochrome P450, family 2, subfamily a, polypeptide 5 |
2175 |
0.27 |
| chr15_27444899_27445331 | 0.20 |
Gm36899 |
predicted gene, 36899 |
18334 |
0.16 |
| chr16_4586554_4586882 | 0.20 |
Glis2 |
GLIS family zinc finger 2 |
7995 |
0.13 |
| chr10_87020834_87020985 | 0.20 |
Stab2 |
stabilin 2 |
12884 |
0.16 |
| chr17_7415036_7415292 | 0.20 |
Gm6553 |
predicted gene 6553 |
15078 |
0.15 |
| chr7_26781568_26782082 | 0.20 |
Cyp2b19 |
cytochrome P450, family 2, subfamily b, polypeptide 19 |
15270 |
0.15 |
| chr2_118874175_118874326 | 0.20 |
Ivd |
isovaleryl coenzyme A dehydrogenase |
57 |
0.96 |
| chr15_66968663_66969063 | 0.20 |
Gm2895 |
predicted gene 2895 |
460 |
0.61 |
| chr5_3596102_3596444 | 0.19 |
Rbm48 |
RNA binding motif protein 48 |
71 |
0.49 |
| chr5_111509241_111509406 | 0.19 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
72099 |
0.08 |
| chr15_99865164_99865320 | 0.19 |
Lima1 |
LIM domain and actin binding 1 |
9429 |
0.08 |
| chr8_12193585_12193751 | 0.19 |
A230072I06Rik |
RIKEN cDNA A230072I06 gene |
85151 |
0.08 |
| chr2_32661930_32662135 | 0.19 |
Mir1954 |
microRNA 1954 |
9703 |
0.07 |
| chr6_94701073_94701362 | 0.19 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
1059 |
0.59 |
| chr12_85350868_85351019 | 0.19 |
Tmed10 |
transmembrane p24 trafficking protein 10 |
23 |
0.96 |
| chr5_139803654_139803817 | 0.19 |
Tmem184a |
transmembrane protein 184a |
4245 |
0.14 |
| chr4_131041255_131041582 | 0.19 |
A930031H19Rik |
RIKEN cDNA A930031H19 gene |
30944 |
0.18 |
| chr18_60748821_60749236 | 0.19 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
378 |
0.77 |
| chr11_16832225_16832426 | 0.19 |
Egfros |
epidermal growth factor receptor, opposite strand |
1623 |
0.42 |
| chr1_45856082_45856244 | 0.19 |
Gm5526 |
predicted pseudogene 5526 |
1591 |
0.29 |
| chr1_160904144_160904325 | 0.19 |
Rc3h1 |
RING CCCH (C3H) domains 1 |
2184 |
0.15 |
| chr1_46854072_46854237 | 0.19 |
Slc39a10 |
solute carrier family 39 (zinc transporter), member 10 |
108 |
0.96 |
| chr10_81320539_81320949 | 0.19 |
Cactin |
cactin, spliceosome C complex subunit |
359 |
0.64 |
| chr5_28079954_28080114 | 0.18 |
Insig1 |
insulin induced gene 1 |
5368 |
0.18 |
| chr11_78697801_78698141 | 0.18 |
Nlk |
nemo like kinase |
598 |
0.68 |
| chr11_16859692_16859843 | 0.18 |
Egfr |
epidermal growth factor receptor |
18383 |
0.18 |
| chr9_63698782_63698955 | 0.18 |
Smad3 |
SMAD family member 3 |
13101 |
0.22 |
| chr3_144570308_144570468 | 0.18 |
Selenof |
selenoprotein F |
38 |
0.72 |
| chrX_164419325_164419506 | 0.18 |
Piga |
phosphatidylinositol glycan anchor biosynthesis, class A |
372 |
0.85 |
| chr7_130631856_130632010 | 0.18 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
13548 |
0.17 |
| chr7_25267913_25268075 | 0.18 |
Cic |
capicua transcriptional repressor |
290 |
0.79 |
| chr18_33437665_33437816 | 0.18 |
Nrep |
neuronal regeneration related protein |
25695 |
0.18 |
| chr8_35077237_35077419 | 0.17 |
Gm34419 |
predicted gene, 34419 |
17217 |
0.16 |
| chr5_73256517_73256913 | 0.17 |
Fryl |
FRY like transcription coactivator |
96 |
0.94 |
| chr8_69774901_69775060 | 0.17 |
Zfp866 |
zinc finger protein 866 |
69 |
0.52 |
| chr2_63862186_63862358 | 0.17 |
Fign |
fidgetin |
235716 |
0.02 |
| chr6_48444394_48444549 | 0.17 |
Zfp467 |
zinc finger protein 467 |
435 |
0.7 |
| chr17_47988159_47988330 | 0.17 |
Gm14871 |
predicted gene 14871 |
15328 |
0.13 |
| chr9_78528236_78528545 | 0.17 |
Gm47430 |
predicted gene, 47430 |
7844 |
0.14 |
| chr14_25456911_25457241 | 0.17 |
Zmiz1os1 |
Zmiz1 opposite strand 1 |
722 |
0.52 |
| chr1_169659535_169659755 | 0.17 |
Rgs5 |
regulator of G-protein signaling 5 |
4123 |
0.26 |
| chr5_75143316_75143477 | 0.17 |
Olfr1401-ps1 |
olfactory receptor 1401, pseudogene 1 |
1838 |
0.21 |
| chr11_53818731_53818912 | 0.17 |
Gm12216 |
predicted gene 12216 |
22946 |
0.11 |
| chr6_127152476_127152648 | 0.17 |
Ccnd2 |
cyclin D2 |
369 |
0.76 |
| chr1_89107539_89107745 | 0.17 |
Gm38312 |
predicted gene, 38312 |
10569 |
0.21 |
| chr6_125165542_125165892 | 0.17 |
Gapdh |
glyceraldehyde-3-phosphate dehydrogenase |
56 |
0.92 |
| chr7_130098725_130098937 | 0.16 |
Gm23847 |
predicted gene, 23847 |
63737 |
0.12 |
| chr16_38376538_38376695 | 0.16 |
Popdc2 |
popeye domain containing 2 |
4618 |
0.14 |
| chr3_88425144_88425305 | 0.16 |
Slc25a44 |
solute carrier family 25, member 44 |
85 |
0.91 |
| chr1_168380327_168380548 | 0.16 |
Gm38381 |
predicted gene, 38381 |
29629 |
0.18 |
| chrX_163958648_163958821 | 0.16 |
Zrsr2 |
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 2 |
73 |
0.98 |
| chr4_57675400_57675766 | 0.16 |
Pakap |
paralemmin A kinase anchor protein |
37608 |
0.18 |
| chr13_72016997_72017161 | 0.16 |
Irx1 |
Iroquois homeobox 1 |
53356 |
0.15 |
| chr13_99794055_99794213 | 0.16 |
Gm24471 |
predicted gene, 24471 |
101605 |
0.06 |
| chr16_43303328_43303503 | 0.16 |
Gm37946 |
predicted gene, 37946 |
4866 |
0.21 |
| chr8_45626757_45627016 | 0.16 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
853 |
0.61 |
| chr1_184732600_184732772 | 0.16 |
Hlx |
H2.0-like homeobox |
67 |
0.96 |
| chr8_124642015_124642295 | 0.16 |
2310022B05Rik |
RIKEN cDNA 2310022B05 gene |
21214 |
0.14 |
| chr2_93261940_93262145 | 0.16 |
Tspan18 |
tetraspanin 18 |
34256 |
0.17 |
| chr13_34814605_34814934 | 0.16 |
Gm47157 |
predicted gene, 47157 |
225 |
0.91 |
| chr17_57214462_57214630 | 0.16 |
Mir6978 |
microRNA 6978 |
2682 |
0.16 |
| chr9_107650834_107650985 | 0.16 |
Slc38a3 |
solute carrier family 38, member 3 |
5111 |
0.08 |
| chr17_7386060_7386243 | 0.16 |
Unc93a2 |
unc-93 homolog A2 |
687 |
0.61 |
| chr16_28851482_28851645 | 0.16 |
Mb21d2 |
Mab-21 domain containing 2 |
15196 |
0.27 |
| chr2_26445181_26445721 | 0.16 |
Sec16a |
SEC16 homolog A, endoplasmic reticulum export factor |
235 |
0.47 |
| chr11_59804844_59804999 | 0.16 |
Flcn |
folliculin |
4667 |
0.11 |
| chr1_134745790_134746030 | 0.16 |
Syt2 |
synaptotagmin II |
2667 |
0.23 |
| chr11_82035273_82035424 | 0.16 |
Ccl2 |
chemokine (C-C motif) ligand 2 |
223 |
0.89 |
| chr10_127668046_127668306 | 0.16 |
Nab2 |
Ngfi-A binding protein 2 |
392 |
0.62 |
| chr1_91448793_91448944 | 0.16 |
Per2 |
period circadian clock 2 |
997 |
0.4 |
| chr12_86478452_86478665 | 0.16 |
Esrrb |
estrogen related receptor, beta |
8441 |
0.26 |
| chr7_141458231_141458600 | 0.16 |
Pnpla2 |
patatin-like phospholipase domain containing 2 |
1397 |
0.16 |
| chr3_68493989_68494284 | 0.16 |
Schip1 |
schwannomin interacting protein 1 |
72 |
0.98 |
| chr3_131564564_131564861 | 0.16 |
Papss1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
56 |
0.98 |
| chr5_150911743_150911914 | 0.16 |
Gm43298 |
predicted gene 43298 |
29186 |
0.16 |
| chr18_49832250_49832445 | 0.16 |
Dmxl1 |
Dmx-like 1 |
323 |
0.9 |
| chr1_132388072_132388223 | 0.15 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
2171 |
0.22 |
| chr5_139730884_139731070 | 0.15 |
Micall2 |
MICAL-like 2 |
1857 |
0.27 |
| chr2_131233930_131234100 | 0.15 |
Mavs |
mitochondrial antiviral signaling protein |
48 |
0.96 |
| chr5_123135984_123136315 | 0.15 |
Gm38102 |
predicted gene, 38102 |
388 |
0.65 |
| chr11_51619519_51619707 | 0.15 |
Nhp2 |
NHP2 ribonucleoprotein |
122 |
0.94 |
| chr19_4240260_4240411 | 0.15 |
Pold4 |
polymerase (DNA-directed), delta 4 |
7897 |
0.06 |
| chr8_57487862_57488025 | 0.15 |
Sap30 |
sin3 associated polypeptide |
83 |
0.53 |
| chr8_89279548_89279868 | 0.15 |
Gm5356 |
predicted pseudogene 5356 |
92148 |
0.09 |
| chr5_134985683_134985851 | 0.15 |
Cldn3 |
claudin 3 |
447 |
0.63 |
| chr12_102788080_102788385 | 0.15 |
Gm47042 |
predicted gene, 47042 |
23520 |
0.08 |
| chr7_139388680_139389161 | 0.15 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
189 |
0.96 |
| chr19_4719341_4719664 | 0.15 |
Sptbn2 |
spectrin beta, non-erythrocytic 2 |
1577 |
0.24 |
| chr2_5951260_5951429 | 0.15 |
Upf2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
125 |
0.96 |
| chr11_87748776_87748945 | 0.15 |
Mir142hg |
Mir142 host gene (non-protein coding) |
6717 |
0.09 |
| chr5_147017350_147017528 | 0.15 |
Lnx2 |
ligand of numb-protein X 2 |
1596 |
0.37 |
| chr19_22304434_22304587 | 0.15 |
Gm22506 |
predicted gene, 22506 |
68892 |
0.12 |
| chr6_29347943_29348124 | 0.15 |
Calu |
calumenin |
36 |
0.96 |
| chr5_122158077_122158391 | 0.15 |
Ppp1cc |
protein phosphatase 1 catalytic subunit gamma |
44 |
0.96 |
| chr2_165326498_165326660 | 0.15 |
Elmo2 |
engulfment and cell motility 2 |
100 |
0.96 |
| chr3_37158282_37158456 | 0.15 |
Gm12532 |
predicted gene 12532 |
24516 |
0.11 |
| chr11_45800072_45800231 | 0.15 |
F630206G17Rik |
RIKEN cDNA F630206G17 gene |
7936 |
0.16 |
| chr10_80139371_80139556 | 0.15 |
Cbarp |
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
98 |
0.88 |
| chr4_136035578_136036045 | 0.14 |
Rpl11 |
ribosomal protein L11 |
7013 |
0.1 |
| chr13_101746090_101746267 | 0.14 |
Gm36638 |
predicted gene, 36638 |
5710 |
0.23 |
| chr3_88296917_88297081 | 0.14 |
Tsacc |
TSSK6 activating co-chaperone |
0 |
0.59 |
| chr4_149854310_149854480 | 0.14 |
Gm47301 |
predicted gene, 47301 |
36631 |
0.09 |
| chr19_57360887_57361068 | 0.14 |
Fam160b1 |
family with sequence similarity 160, member B1 |
53 |
0.7 |
| chr19_36731141_36731447 | 0.14 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
5359 |
0.23 |
| chr16_20716658_20716809 | 0.14 |
Clcn2 |
chloride channel, voltage-sensitive 2 |
569 |
0.44 |
| chr18_58481759_58481942 | 0.14 |
Slc27a6 |
solute carrier family 27 (fatty acid transporter), member 6 |
74407 |
0.12 |
| chr1_171344047_171344210 | 0.14 |
Nit1 |
nitrilase 1 |
339 |
0.66 |
| chr6_108065232_108065404 | 0.14 |
Setmar |
SET domain without mariner transposase fusion |
256 |
0.94 |
| chr10_128337509_128337866 | 0.14 |
Cs |
citrate synthase |
47 |
0.92 |
| chr12_84772846_84773376 | 0.14 |
Npc2 |
NPC intracellular cholesterol transporter 2 |
41 |
0.62 |
| chr3_89391856_89392032 | 0.14 |
Gm15417 |
predicted gene 15417 |
26 |
0.6 |
| chr13_67060869_67061048 | 0.14 |
Gm46440 |
predicted gene, 46440 |
157 |
0.55 |
| chr4_123411950_123412127 | 0.14 |
Macf1 |
microtubule-actin crosslinking factor 1 |
190 |
0.94 |
| chr1_39950143_39950310 | 0.14 |
Map4k4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
13929 |
0.22 |
| chr1_168432437_168432592 | 0.14 |
Pbx1 |
pre B cell leukemia homeobox 1 |
244 |
0.95 |
| chr11_69996851_69997024 | 0.14 |
Phf23 |
PHD finger protein 23 |
4 |
0.92 |
| chr11_51494380_51494743 | 0.14 |
Col23a1 |
collagen, type XXIII, alpha 1 |
79871 |
0.07 |
| chr6_124481749_124481909 | 0.14 |
C1rl |
complement component 1, r subcomponent-like |
11284 |
0.09 |
| chr9_72767021_72767358 | 0.14 |
Gm27204 |
predicted gene 27204 |
9036 |
0.12 |
| chr11_106512401_106512774 | 0.14 |
Gm22711 |
predicted gene, 22711 |
11342 |
0.15 |
| chr10_77864430_77864592 | 0.14 |
Tspear |
thrombospondin type laminin G domain and EAR repeats |
71 |
0.91 |
| chr10_88507811_88508139 | 0.14 |
Chpt1 |
choline phosphotransferase 1 |
3902 |
0.18 |
| chr5_113024342_113024526 | 0.14 |
Grk3 |
G protein-coupled receptor kinase 3 |
8643 |
0.14 |
| chr8_34804694_34804863 | 0.14 |
Dusp4 |
dual specificity phosphatase 4 |
2519 |
0.32 |
| chr6_94696254_94696442 | 0.14 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
3789 |
0.28 |
| chr5_118332124_118332291 | 0.14 |
Gm28563 |
predicted gene 28563 |
65123 |
0.09 |
| chr2_105396213_105396364 | 0.14 |
Rcn1 |
reticulocalbin 1 |
940 |
0.67 |
| chr2_73003161_73003312 | 0.14 |
Sp3os |
trans-acting transcription factor 3, opposite strand |
15995 |
0.14 |
| chr7_28938830_28938981 | 0.14 |
Gm44699 |
predicted gene 44699 |
4623 |
0.12 |
| chr5_24941855_24942029 | 0.14 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
32965 |
0.13 |
| chr11_40694837_40695013 | 0.13 |
Mat2b |
methionine adenosyltransferase II, beta |
62 |
0.97 |
| chr13_55211103_55211935 | 0.13 |
Nsd1 |
nuclear receptor-binding SET-domain protein 1 |
22 |
0.97 |
| chr5_23890845_23891003 | 0.13 |
AC117663.1 |
solute carrier family 40 (iron-regulated transporter), member 1 (Slc40a1), pseudogene |
14481 |
0.11 |
| chr9_45268129_45268280 | 0.13 |
Il10ra |
interleukin 10 receptor, alpha |
942 |
0.35 |
| chr5_137339583_137339751 | 0.13 |
Gm43012 |
predicted gene 43012 |
2258 |
0.14 |
| chr2_158168887_158169091 | 0.13 |
Tgm2 |
transglutaminase 2, C polypeptide |
22553 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.5 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.2 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.0 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.1 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.1 | GO:0072262 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0060534 | trachea cartilage development(GO:0060534) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.0 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.3 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.3 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.0 | GO:0034893 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |