Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
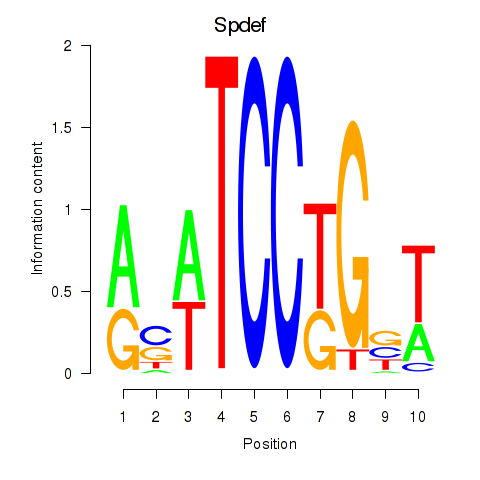
Results for Spdef
Z-value: 1.07
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSMUSG00000024215.7 | SAM pointed domain containing ets transcription factor |
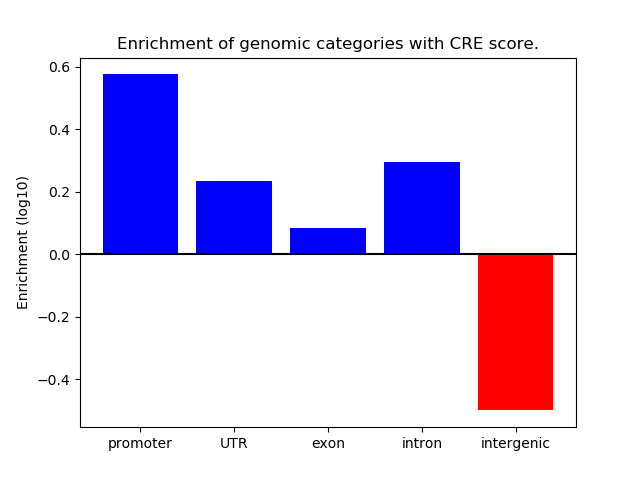
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_27716973_27717159 | Spdef | 653 | 0.588600 | -0.89 | 1.7e-02 | Click! |
| chr17_27716769_27716920 | Spdef | 875 | 0.465415 | -0.65 | 1.6e-01 | Click! |
| chr17_27721382_27721552 | Spdef | 3118 | 0.160414 | 0.35 | 4.9e-01 | Click! |
| chr17_27717560_27717711 | Spdef | 84 | 0.952160 | -0.31 | 5.5e-01 | Click! |
Activity of the Spdef motif across conditions
Conditions sorted by the z-value of the Spdef motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
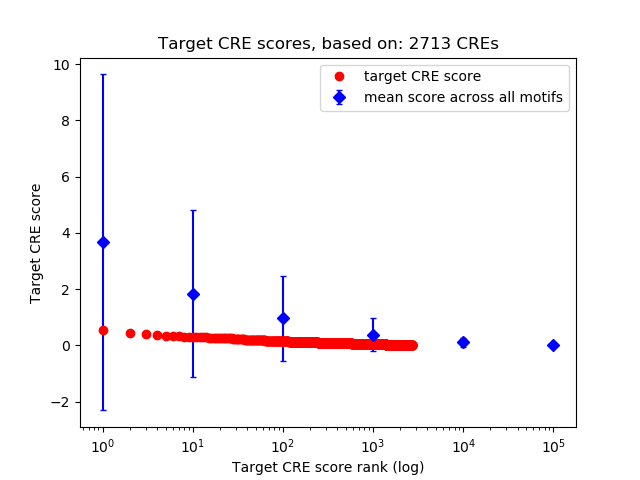
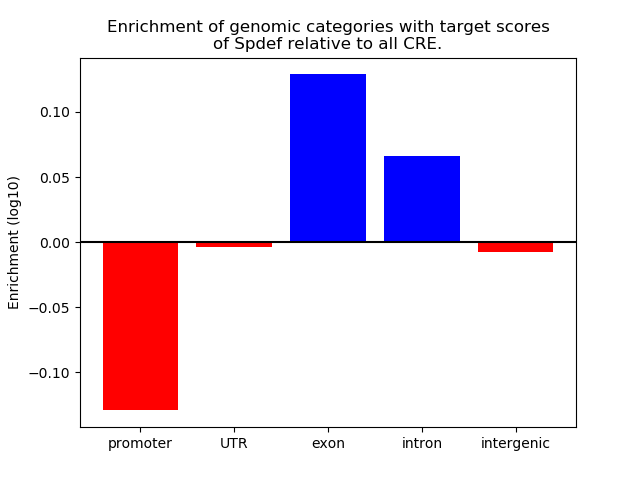
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_97413837_97414007 | 0.55 |
Thrsp |
thyroid hormone responsive |
3597 |
0.16 |
| chr12_4220651_4220920 | 0.44 |
Gm48210 |
predicted gene, 48210 |
1317 |
0.27 |
| chr17_84226805_84226999 | 0.42 |
Thada |
thyroid adenoma associated |
2958 |
0.24 |
| chr16_45377470_45377621 | 0.37 |
Cd200 |
CD200 antigen |
22767 |
0.16 |
| chr1_67134415_67135075 | 0.34 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
11719 |
0.24 |
| chr11_16844975_16845160 | 0.33 |
Egfros |
epidermal growth factor receptor, opposite strand |
14365 |
0.2 |
| chr17_8797635_8797786 | 0.32 |
Pde10a |
phosphodiesterase 10A |
3983 |
0.29 |
| chr4_63355986_63356174 | 0.31 |
Orm3 |
orosomucoid 3 |
82 |
0.95 |
| chr14_28998848_28998999 | 0.30 |
Gm23999 |
predicted gene, 23999 |
10876 |
0.19 |
| chr6_88003582_88003770 | 0.30 |
4933412L11Rik |
RIKEN cDNA 4933412L11 gene |
9497 |
0.09 |
| chr14_52012195_52012346 | 0.29 |
Zfp219 |
zinc finger protein 219 |
1388 |
0.21 |
| chr5_43146388_43146587 | 0.29 |
Gm42552 |
predicted gene 42552 |
1840 |
0.38 |
| chr3_69280240_69280391 | 0.29 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
36546 |
0.13 |
| chr3_31970158_31970309 | 0.28 |
Gm37834 |
predicted gene, 37834 |
15469 |
0.27 |
| chr12_4220183_4220334 | 0.27 |
Gm48210 |
predicted gene, 48210 |
1844 |
0.19 |
| chr9_122849425_122849576 | 0.27 |
Gm47140 |
predicted gene, 47140 |
1082 |
0.34 |
| chr2_72741921_72742104 | 0.27 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
8841 |
0.22 |
| chr12_103991999_103992602 | 0.27 |
Serpina11 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
2343 |
0.16 |
| chr5_50031875_50032113 | 0.26 |
Adgra3 |
adhesion G protein-coupled receptor A3 |
21385 |
0.19 |
| chr9_46092730_46092881 | 0.26 |
Sik3 |
SIK family kinase 3 |
30325 |
0.14 |
| chr16_43330685_43330836 | 0.25 |
Gm15711 |
predicted gene 15711 |
18166 |
0.16 |
| chr6_146751805_146751963 | 0.25 |
Gm44270 |
predicted gene, 44270 |
10697 |
0.14 |
| chr4_63350215_63350540 | 0.25 |
Gm11212 |
predicted gene 11212 |
34 |
0.96 |
| chr8_76148232_76148470 | 0.25 |
Gm45742 |
predicted gene 45742 |
31324 |
0.21 |
| chr7_35748679_35748830 | 0.25 |
Dpy19l3 |
dpy-19-like 3 (C. elegans) |
5601 |
0.2 |
| chr19_11977402_11977711 | 0.25 |
Osbp |
oxysterol binding protein |
11615 |
0.09 |
| chr6_137773601_137773762 | 0.24 |
Dera |
deoxyribose-phosphate aldolase (putative) |
1308 |
0.52 |
| chr3_94702452_94702603 | 0.24 |
Selenbp2 |
selenium binding protein 2 |
8868 |
0.11 |
| chr10_40502877_40503043 | 0.22 |
Gm18671 |
predicted gene, 18671 |
45932 |
0.12 |
| chr6_38144404_38144577 | 0.22 |
D630045J12Rik |
RIKEN cDNA D630045J12 gene |
2884 |
0.23 |
| chr19_44399974_44400175 | 0.22 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6616 |
0.15 |
| chr5_125040097_125040498 | 0.22 |
Ncor2 |
nuclear receptor co-repressor 2 |
2515 |
0.25 |
| chr1_71735697_71735877 | 0.22 |
Gm5256 |
predicted gene 5256 |
20863 |
0.17 |
| chr3_97635501_97635936 | 0.21 |
Fmo5 |
flavin containing monooxygenase 5 |
6835 |
0.14 |
| chr1_88248965_88249116 | 0.21 |
Mroh2a |
maestro heat-like repeat family member 2A |
8718 |
0.09 |
| chr11_120810230_120810550 | 0.21 |
Fasn |
fatty acid synthase |
689 |
0.48 |
| chr16_43234125_43234650 | 0.21 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1493 |
0.45 |
| chr17_35254998_35255149 | 0.21 |
Mir8094 |
microRNA 8094 |
3784 |
0.06 |
| chr12_104088559_104088873 | 0.20 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
8067 |
0.1 |
| chr11_119007787_119007988 | 0.19 |
Cbx2 |
chromobox 2 |
15075 |
0.15 |
| chr19_25422520_25422717 | 0.19 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
379 |
0.88 |
| chr9_74228469_74228620 | 0.19 |
Wdr72 |
WD repeat domain 72 |
45590 |
0.18 |
| chr2_112595326_112595515 | 0.19 |
Gm44374 |
predicted gene, 44374 |
18610 |
0.17 |
| chr11_16791951_16792316 | 0.19 |
Egfros |
epidermal growth factor receptor, opposite strand |
38569 |
0.14 |
| chr13_101718863_101719071 | 0.19 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
16863 |
0.2 |
| chr5_122320103_122320299 | 0.19 |
Gm15842 |
predicted gene 15842 |
17779 |
0.09 |
| chr18_12692319_12692517 | 0.19 |
Ttc39c |
tetratricopeptide repeat domain 39C |
2690 |
0.21 |
| chr2_58761406_58761587 | 0.18 |
Upp2 |
uridine phosphorylase 2 |
3829 |
0.24 |
| chr4_154297797_154298168 | 0.18 |
Arhgef16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
1910 |
0.29 |
| chr19_34270258_34270414 | 0.18 |
Gm50239 |
predicted gene, 50239 |
14201 |
0.14 |
| chr10_42708978_42709129 | 0.18 |
Gm15200 |
predicted gene 15200 |
10535 |
0.18 |
| chr7_123643560_123643752 | 0.18 |
Zkscan2 |
zinc finger with KRAB and SCAN domains 2 |
143207 |
0.04 |
| chr3_94705046_94705225 | 0.18 |
Selenbp2 |
selenium binding protein 2 |
11476 |
0.11 |
| chr2_58761173_58761336 | 0.18 |
Upp2 |
uridine phosphorylase 2 |
4071 |
0.24 |
| chr12_104740709_104740870 | 0.18 |
Dicer1 |
dicer 1, ribonuclease type III |
1952 |
0.38 |
| chr17_28016505_28016679 | 0.18 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
9247 |
0.12 |
| chr11_60196823_60197490 | 0.18 |
Mir6921 |
microRNA 6921 |
3461 |
0.14 |
| chr6_24046716_24047018 | 0.17 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
61704 |
0.12 |
| chr10_128206622_128206773 | 0.17 |
Gls2 |
glutaminase 2 (liver, mitochondrial) |
228 |
0.82 |
| chr11_5906758_5906909 | 0.17 |
Gck |
glucokinase |
4746 |
0.13 |
| chr10_4731451_4731623 | 0.17 |
Esr1 |
estrogen receptor 1 (alpha) |
18886 |
0.26 |
| chr11_60202909_60203232 | 0.17 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
418 |
0.59 |
| chr2_30144800_30144973 | 0.17 |
Tbc1d13 |
TBC1 domain family, member 13 |
2607 |
0.15 |
| chr9_121881807_121881992 | 0.17 |
Gm47113 |
predicted gene, 47113 |
12403 |
0.08 |
| chr3_21986159_21986455 | 0.17 |
Gm43674 |
predicted gene 43674 |
12161 |
0.21 |
| chr8_104831387_104831616 | 0.17 |
Ces2b |
carboxyesterase 2B |
71 |
0.94 |
| chr5_87562697_87562848 | 0.17 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
2581 |
0.17 |
| chr7_72260804_72260982 | 0.17 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
31533 |
0.22 |
| chr12_70720580_70720731 | 0.17 |
Gm32369 |
predicted gene, 32369 |
38805 |
0.12 |
| chr2_91750560_91750711 | 0.16 |
Ambra1 |
autophagy/beclin 1 regulator 1 |
15810 |
0.14 |
| chr11_60816993_60817396 | 0.16 |
Gm12611 |
predicted gene 12611 |
819 |
0.38 |
| chr19_44397890_44398071 | 0.16 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
8710 |
0.15 |
| chr18_12711826_12711991 | 0.16 |
Mir1948 |
microRNA 1948 |
2903 |
0.2 |
| chr12_86826629_86826835 | 0.16 |
Gm10095 |
predicted gene 10095 |
19735 |
0.17 |
| chr9_21838672_21838867 | 0.16 |
Angptl8 |
angiopoietin-like 8 |
3259 |
0.14 |
| chr4_63348935_63349236 | 0.16 |
Gm11212 |
predicted gene 11212 |
1326 |
0.29 |
| chr2_181522503_181522654 | 0.16 |
Dnajc5 |
DnaJ heat shock protein family (Hsp40) member C5 |
1631 |
0.22 |
| chr10_58228380_58228585 | 0.16 |
Gm10807 |
predicted gene 10807 |
169 |
0.92 |
| chr10_99171877_99172060 | 0.16 |
Poc1b |
POC1 centriolar protein B |
684 |
0.6 |
| chr7_116183557_116183712 | 0.16 |
Plekha7 |
pleckstrin homology domain containing, family A member 7 |
881 |
0.63 |
| chr12_75448996_75449191 | 0.16 |
Gphb5 |
glycoprotein hormone beta 5 |
32312 |
0.18 |
| chr17_46073702_46073982 | 0.16 |
Gm36200 |
predicted gene, 36200 |
10424 |
0.13 |
| chr8_82255910_82256078 | 0.16 |
Gm38590 |
predicted gene, 38590 |
48459 |
0.15 |
| chr14_65615417_65615662 | 0.15 |
Nuggc |
nuclear GTPase, germinal center associated |
10272 |
0.2 |
| chr9_116185819_116186009 | 0.15 |
Gm4668 |
predicted gene 4668 |
3266 |
0.22 |
| chr4_152264194_152264355 | 0.15 |
Gpr153 |
G protein-coupled receptor 153 |
9958 |
0.12 |
| chr5_90468289_90468670 | 0.15 |
Alb |
albumin |
5779 |
0.16 |
| chr3_27704926_27705320 | 0.15 |
Fndc3b |
fibronectin type III domain containing 3B |
5275 |
0.31 |
| chr6_97962459_97962619 | 0.15 |
Mitf |
melanogenesis associated transcription factor |
21621 |
0.24 |
| chr13_23298661_23298830 | 0.15 |
Gm11333 |
predicted gene 11333 |
696 |
0.46 |
| chr10_114191151_114191309 | 0.15 |
Gm48104 |
predicted gene, 48104 |
95503 |
0.09 |
| chr4_32290238_32290389 | 0.15 |
Bach2it1 |
BTB and CNC homology 2, intronic transcript 1 |
38552 |
0.16 |
| chr13_108338110_108338518 | 0.15 |
Depdc1b |
DEP domain containing 1B |
11786 |
0.18 |
| chr4_45800098_45800462 | 0.15 |
Aldh1b1 |
aldehyde dehydrogenase 1 family, member B1 |
1159 |
0.41 |
| chr14_17768072_17768293 | 0.15 |
Gm48320 |
predicted gene, 48320 |
2940 |
0.39 |
| chr2_156722367_156722542 | 0.14 |
Dlgap4 |
DLG associated protein 4 |
962 |
0.41 |
| chr10_8230404_8230563 | 0.14 |
Gm30906 |
predicted gene, 30906 |
54352 |
0.15 |
| chr8_34164335_34164526 | 0.14 |
Mir6395 |
microRNA 6395 |
2282 |
0.18 |
| chr14_21181187_21181360 | 0.14 |
Adk |
adenosine kinase |
105121 |
0.06 |
| chr4_97885463_97885690 | 0.14 |
Nfia |
nuclear factor I/A |
5457 |
0.31 |
| chr6_28423868_28424057 | 0.14 |
Gcc1 |
golgi coiled coil 1 |
191 |
0.56 |
| chr7_28957656_28957827 | 0.14 |
Actn4 |
actinin alpha 4 |
4482 |
0.12 |
| chr8_84723903_84724146 | 0.14 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
1017 |
0.36 |
| chr11_16880492_16880986 | 0.14 |
Egfr |
epidermal growth factor receptor |
2589 |
0.29 |
| chr9_120133572_120133723 | 0.14 |
Gm5922 |
predicted gene 5922 |
1368 |
0.2 |
| chr7_106236793_106236971 | 0.14 |
Gvin1 |
GTPase, very large interferon inducible 1 |
21556 |
0.16 |
| chr4_72398261_72398417 | 0.14 |
Gm11235 |
predicted gene 11235 |
144327 |
0.05 |
| chr11_16816779_16816940 | 0.14 |
Egfros |
epidermal growth factor receptor, opposite strand |
13843 |
0.21 |
| chr17_43242727_43242878 | 0.14 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
27527 |
0.22 |
| chr17_86552135_86552300 | 0.14 |
Gm10309 |
predicted gene 10309 |
46985 |
0.15 |
| chr1_153862795_153862997 | 0.14 |
Teddm1b |
transmembrane epididymal protein 1B |
11449 |
0.1 |
| chr1_186684187_186684350 | 0.14 |
Gm26169 |
predicted gene, 26169 |
7526 |
0.16 |
| chr12_103947943_103948118 | 0.14 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
8868 |
0.11 |
| chr6_87913437_87913823 | 0.14 |
Hmces |
5-hydroxymethylcytosine (hmC) binding, ES cell specific |
305 |
0.71 |
| chr7_16405904_16406273 | 0.14 |
Gm45508 |
predicted gene 45508 |
1826 |
0.18 |
| chr13_50047188_50047373 | 0.13 |
Gm48052 |
predicted gene, 48052 |
3725 |
0.16 |
| chr3_18107731_18107882 | 0.13 |
Gm23726 |
predicted gene, 23726 |
38480 |
0.15 |
| chr7_139416534_139416685 | 0.13 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
16120 |
0.23 |
| chr11_102496845_102497026 | 0.13 |
Gm11628 |
predicted gene 11628 |
4326 |
0.12 |
| chr14_27185973_27186174 | 0.13 |
Gm7591 |
predicted gene 7591 |
24324 |
0.18 |
| chr15_79414701_79414859 | 0.13 |
Csnk1e |
casein kinase 1, epsilon |
6535 |
0.11 |
| chr9_56938509_56938699 | 0.13 |
Imp3 |
IMP3, U3 small nucleolar ribonucleoprotein |
1129 |
0.32 |
| chr2_178606624_178606775 | 0.13 |
Cdh26 |
cadherin-like 26 |
146069 |
0.04 |
| chr15_85868388_85868539 | 0.13 |
Gtse1 |
G two S phase expressed protein 1 |
8517 |
0.12 |
| chr13_23307496_23307808 | 0.13 |
4930557F10Rik |
RIKEN cDNA 4930557F10 gene |
5748 |
0.09 |
| chr8_72169355_72169641 | 0.13 |
Rab8a |
RAB8A, member RAS oncogene family |
8139 |
0.09 |
| chr6_38120784_38120952 | 0.13 |
Gm23231 |
predicted gene, 23231 |
2485 |
0.22 |
| chr10_117906642_117906793 | 0.13 |
4933411E08Rik |
RIKEN cDNA 4933411E08 gene |
18742 |
0.15 |
| chr18_69537508_69537694 | 0.13 |
Gm24845 |
predicted gene, 24845 |
14362 |
0.23 |
| chr2_27381428_27381611 | 0.13 |
Gm24049 |
predicted gene, 24049 |
20333 |
0.14 |
| chr11_44546590_44546755 | 0.13 |
Rnf145 |
ring finger protein 145 |
10159 |
0.18 |
| chr3_94398765_94399136 | 0.13 |
Lingo4 |
leucine rich repeat and Ig domain containing 4 |
433 |
0.59 |
| chr2_58769522_58769673 | 0.13 |
Upp2 |
uridine phosphorylase 2 |
4272 |
0.24 |
| chr3_21905574_21905725 | 0.13 |
Gm43674 |
predicted gene 43674 |
92819 |
0.08 |
| chr12_52505478_52505629 | 0.13 |
Arhgap5 |
Rho GTPase activating protein 5 |
1211 |
0.47 |
| chr5_20319781_20319982 | 0.13 |
Gm22755 |
predicted gene, 22755 |
55540 |
0.13 |
| chr5_125490850_125491058 | 0.13 |
Gm27551 |
predicted gene, 27551 |
11577 |
0.13 |
| chr17_26538205_26538356 | 0.12 |
Gm50275 |
predicted gene, 50275 |
53 |
0.95 |
| chr6_90491129_90491426 | 0.12 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
4850 |
0.12 |
| chrX_75673853_75674412 | 0.12 |
Gm15065 |
predicted gene 15065 |
31277 |
0.13 |
| chr1_67207883_67208119 | 0.12 |
Gm15668 |
predicted gene 15668 |
41199 |
0.15 |
| chr9_118099534_118099709 | 0.12 |
Azi2 |
5-azacytidine induced gene 2 |
40565 |
0.13 |
| chr10_79879613_79879900 | 0.12 |
Prtn3 |
proteinase 3 |
142 |
0.85 |
| chr10_99169914_99170080 | 0.12 |
Poc1b |
POC1 centriolar protein B |
1287 |
0.35 |
| chr13_9097997_9098273 | 0.12 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
4153 |
0.18 |
| chr12_112630609_112630843 | 0.12 |
Adssl1 |
adenylosuccinate synthetase like 1 |
8646 |
0.11 |
| chr8_105235292_105235443 | 0.12 |
D230025D16Rik |
RIKEN cDNA D230025D16 gene |
4399 |
0.09 |
| chr11_72472528_72472733 | 0.12 |
Spns2 |
spinster homolog 2 |
2397 |
0.21 |
| chr9_107544674_107544842 | 0.12 |
Nprl2 |
NPR2 like, GATOR1 complex subunit |
780 |
0.3 |
| chr11_105974677_105975033 | 0.12 |
Ace |
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
353 |
0.78 |
| chr4_36131695_36131853 | 0.12 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
4689 |
0.35 |
| chr6_94050888_94051087 | 0.12 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
137297 |
0.05 |
| chr6_38122561_38122726 | 0.12 |
Atp6v0a4 |
ATPase, H+ transporting, lysosomal V0 subunit A4 |
1846 |
0.28 |
| chr4_63360320_63360510 | 0.12 |
Orm2 |
orosomucoid 2 |
2034 |
0.19 |
| chr15_82362153_82362313 | 0.12 |
Ndufa6 |
NADH:ubiquinone oxidoreductase subunit A6 |
7911 |
0.07 |
| chr2_59979696_59979870 | 0.12 |
Gm13574 |
predicted gene 13574 |
12561 |
0.19 |
| chr4_35069175_35069344 | 0.12 |
Ifnk |
interferon kappa |
82797 |
0.07 |
| chr16_43381191_43381342 | 0.12 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
17062 |
0.17 |
| chr16_24860524_24860695 | 0.12 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
15434 |
0.2 |
| chr12_103311070_103311221 | 0.12 |
Fam181a |
family with sequence similarity 181, member A |
170 |
0.92 |
| chr15_77548351_77548502 | 0.12 |
1700025B11Rik |
RIKEN cDNA 1700025B11 gene |
13466 |
0.11 |
| chr3_137943821_137943983 | 0.12 |
Dapp1 |
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
10811 |
0.1 |
| chr14_11132816_11133186 | 0.12 |
Fhit |
fragile histidine triad gene |
7410 |
0.24 |
| chr17_70990050_70990412 | 0.12 |
Myl12b |
myosin, light chain 12B, regulatory |
299 |
0.82 |
| chr19_8128072_8128223 | 0.12 |
Slc22a28 |
solute carrier family 22, member 28 |
3835 |
0.25 |
| chr1_67170641_67171204 | 0.12 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
47896 |
0.14 |
| chr14_121720285_121720661 | 0.12 |
Dock9 |
dedicator of cytokinesis 9 |
18388 |
0.23 |
| chr6_96116339_96116711 | 0.12 |
Tafa1 |
TAFA chemokine like family member 1 |
1214 |
0.58 |
| chr17_87379036_87379189 | 0.12 |
Ttc7 |
tetratricopeptide repeat domain 7 |
259 |
0.89 |
| chr11_16819306_16819457 | 0.12 |
Egfros |
epidermal growth factor receptor, opposite strand |
11321 |
0.22 |
| chr2_31745589_31745792 | 0.11 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
14253 |
0.14 |
| chr18_21242831_21243130 | 0.11 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
57143 |
0.11 |
| chr9_72720556_72720835 | 0.11 |
Gm37955 |
predicted gene, 37955 |
3735 |
0.13 |
| chr7_38209598_38209825 | 0.11 |
D530033B14Rik |
RIKEN cDNA D530033B14 gene |
13254 |
0.12 |
| chr14_7841370_7841604 | 0.11 |
Flnb |
filamin, beta |
23530 |
0.15 |
| chr6_48493256_48493431 | 0.11 |
Sspo |
SCO-spondin |
437 |
0.68 |
| chr11_120809505_120809952 | 0.11 |
Fasn |
fatty acid synthase |
27 |
0.95 |
| chr6_90535262_90535425 | 0.11 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
15446 |
0.12 |
| chr4_101331247_101331398 | 0.11 |
Gm12796 |
predicted gene 12796 |
9577 |
0.1 |
| chr10_95146574_95146766 | 0.11 |
Gm29684 |
predicted gene, 29684 |
845 |
0.56 |
| chr9_103249018_103249229 | 0.11 |
1300017J02Rik |
RIKEN cDNA 1300017J02 gene |
7903 |
0.17 |
| chr4_153019323_153019495 | 0.11 |
Gm25779 |
predicted gene, 25779 |
5825 |
0.32 |
| chr8_35208499_35208650 | 0.11 |
Gm34474 |
predicted gene, 34474 |
10064 |
0.14 |
| chr4_122872061_122872212 | 0.11 |
Cap1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
7504 |
0.17 |
| chr7_121043590_121043802 | 0.11 |
Gm27991 |
predicted gene, 27991 |
3216 |
0.11 |
| chr17_34153027_34153195 | 0.11 |
H2-DMb1 |
histocompatibility 2, class II, locus Mb1 |
39 |
0.92 |
| chr14_76149595_76149834 | 0.11 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
38823 |
0.15 |
| chr12_70991810_70992010 | 0.11 |
Psma3 |
proteasome subunit alpha 3 |
5018 |
0.16 |
| chr9_53344725_53344910 | 0.11 |
Exph5 |
exophilin 5 |
3628 |
0.22 |
| chr5_151103258_151103409 | 0.11 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
5402 |
0.28 |
| chr4_108318788_108318978 | 0.11 |
Gm12740 |
predicted gene 12740 |
3455 |
0.16 |
| chr6_39259262_39259413 | 0.11 |
Gm26008 |
predicted gene, 26008 |
6605 |
0.17 |
| chr14_65722029_65722405 | 0.11 |
Scara5 |
scavenger receptor class A, member 5 |
45740 |
0.14 |
| chr9_109123416_109123604 | 0.11 |
Plxnb1 |
plexin B1 |
16716 |
0.1 |
| chr1_92903311_92903462 | 0.11 |
Ankmy1 |
ankyrin repeat and MYND domain containing 1 |
480 |
0.64 |
| chr1_131990500_131990816 | 0.11 |
Slc45a3 |
solute carrier family 45, member 3 |
11694 |
0.12 |
| chr1_67146866_67147049 | 0.11 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
23931 |
0.21 |
| chr14_120362393_120362544 | 0.11 |
Mbnl2 |
muscleblind like splicing factor 2 |
16773 |
0.22 |
| chr5_30643999_30644150 | 0.11 |
Slc35f6 |
solute carrier family 35, member F6 |
3859 |
0.14 |
| chr2_52579266_52579417 | 0.11 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
20774 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.0 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0035276 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0034785 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |