Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
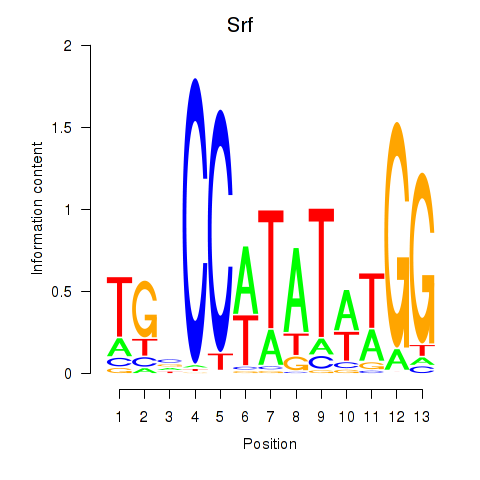
Results for Srf
Z-value: 2.15
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSMUSG00000015605.5 | serum response factor |
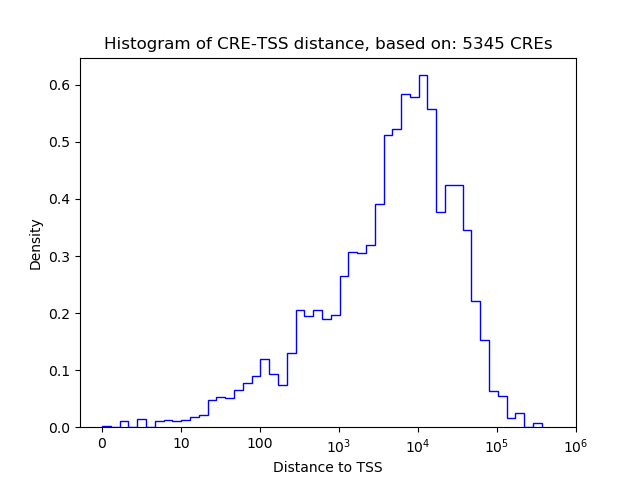
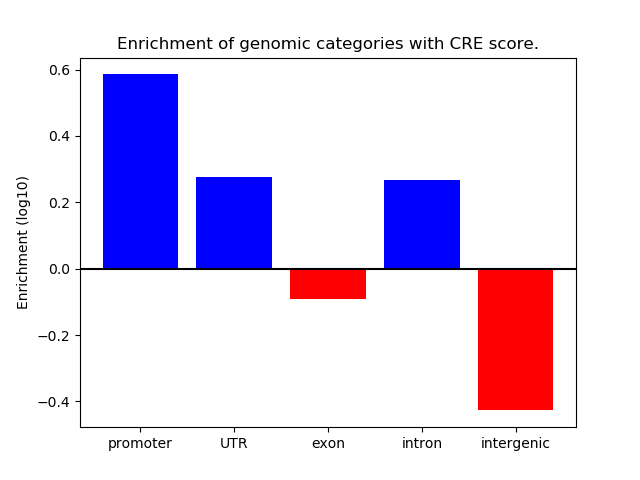
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46556498_46556886 | Srf | 517 | 0.635445 | -0.69 | 1.3e-01 | Click! |
| chr17_46555816_46556119 | Srf | 208 | 0.874547 | -0.52 | 3.0e-01 | Click! |
| chr17_46556258_46556418 | Srf | 163 | 0.905587 | 0.47 | 3.5e-01 | Click! |
Activity of the Srf motif across conditions
Conditions sorted by the z-value of the Srf motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
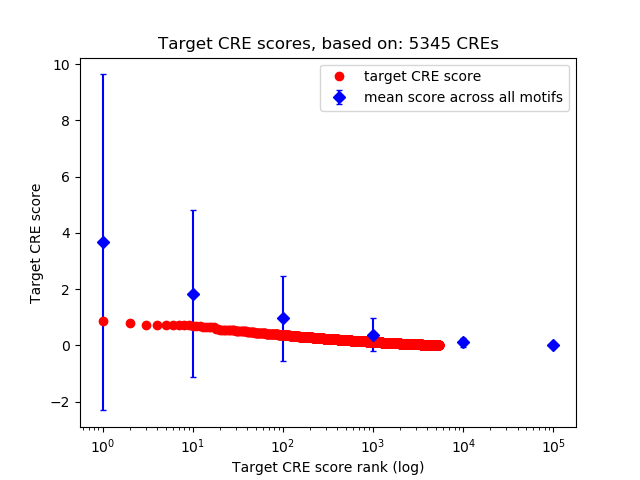
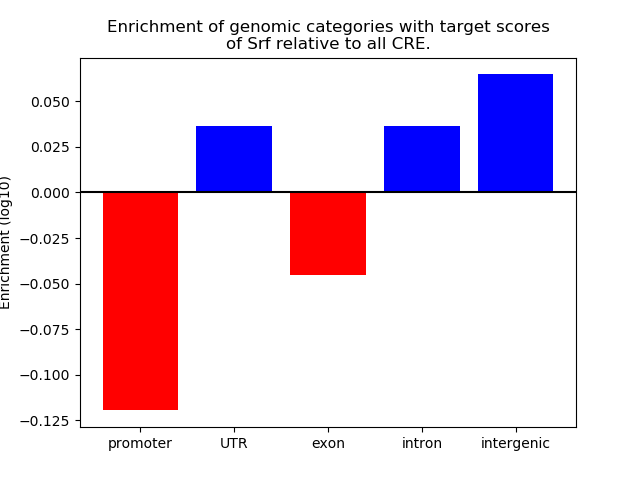
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_16830224_16830396 | 0.87 |
Egfros |
epidermal growth factor receptor, opposite strand |
392 |
0.88 |
| chr3_69139928_69140109 | 0.80 |
Gm1647 |
predicted gene 1647 |
8785 |
0.14 |
| chr7_44473035_44473343 | 0.74 |
5430431A17Rik |
RIKEN cDNA 5430431A17 gene |
349 |
0.67 |
| chr2_72223053_72223218 | 0.74 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
9167 |
0.17 |
| chr15_7135984_7136307 | 0.73 |
Lifr |
LIF receptor alpha |
4397 |
0.32 |
| chr13_59533414_59533566 | 0.73 |
Agtpbp1 |
ATP/GTP binding protein 1 |
300 |
0.88 |
| chr1_184606698_184606874 | 0.72 |
Rpl21-ps1 |
ribosomal protein 21, pseudogene 1 |
10462 |
0.15 |
| chr14_30254871_30255032 | 0.72 |
Gm47570 |
predicted gene, 47570 |
47213 |
0.13 |
| chr14_20892049_20892245 | 0.71 |
Gm6128 |
predicted pseudogene 6128 |
35726 |
0.13 |
| chr7_114290333_114290662 | 0.70 |
Psma1 |
proteasome subunit alpha 1 |
14379 |
0.21 |
| chr16_45806302_45806490 | 0.69 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
4825 |
0.22 |
| chr4_137810112_137810287 | 0.68 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
13815 |
0.19 |
| chr5_120103213_120103364 | 0.67 |
Rbm19 |
RNA binding motif protein 19 |
13177 |
0.19 |
| chr7_106644629_106644780 | 0.66 |
Olfr693 |
olfactory receptor 693 |
33781 |
0.1 |
| chr8_41545072_41545241 | 0.65 |
Gapdh-ps14 |
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 14 |
64513 |
0.13 |
| chr2_118318601_118318758 | 0.64 |
1700054M17Rik |
RIKEN cDNA 1700054M17 gene |
13765 |
0.13 |
| chr18_11851878_11852035 | 0.64 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
7656 |
0.17 |
| chr7_77888814_77888965 | 0.58 |
Gm23239 |
predicted gene, 23239 |
21431 |
0.28 |
| chr10_68248229_68248529 | 0.56 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
30342 |
0.19 |
| chr1_106200916_106201079 | 0.56 |
Gm38235 |
predicted gene, 38235 |
18317 |
0.17 |
| chr12_85462932_85463085 | 0.55 |
Fos |
FBJ osteosarcoma oncogene |
10882 |
0.16 |
| chrX_111648183_111648334 | 0.55 |
Hdx |
highly divergent homeobox |
48821 |
0.16 |
| chr9_49514430_49514581 | 0.55 |
Gm11149 |
predicted gene 11149 |
3783 |
0.24 |
| chr6_136930874_136931029 | 0.54 |
Pde6h |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
7119 |
0.13 |
| chr6_38949943_38950137 | 0.54 |
Tbxas1 |
thromboxane A synthase 1, platelet |
8639 |
0.19 |
| chr5_21139662_21139813 | 0.54 |
Gm42660 |
predicted gene 42660 |
8661 |
0.14 |
| chr3_18146281_18146519 | 0.54 |
Gm23686 |
predicted gene, 23686 |
31225 |
0.18 |
| chr12_110381296_110381622 | 0.53 |
Gm47195 |
predicted gene, 47195 |
56074 |
0.08 |
| chr16_18187372_18187523 | 0.53 |
Mir6366 |
microRNA 6366 |
22277 |
0.08 |
| chr2_122086820_122087004 | 0.52 |
Spg11 |
SPG11, spatacsin vesicle trafficking associated |
31384 |
0.11 |
| chr11_16851040_16851282 | 0.51 |
Egfros |
epidermal growth factor receptor, opposite strand |
20459 |
0.18 |
| chr5_138255290_138255446 | 0.51 |
Lamtor4 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
114 |
0.89 |
| chr5_113872987_113873184 | 0.51 |
Coro1c |
coronin, actin binding protein 1C |
1563 |
0.25 |
| chr2_92380520_92380699 | 0.51 |
1700029I15Rik |
RIKEN cDNA 1700029I15 gene |
1929 |
0.18 |
| chr10_80289084_80289235 | 0.51 |
Rps15 |
ribosomal protein S15 |
3294 |
0.08 |
| chr2_31165165_31165344 | 0.50 |
Gpr107 |
G protein-coupled receptor 107 |
7013 |
0.14 |
| chr11_74493309_74493480 | 0.49 |
Rap1gap2 |
RAP1 GTPase activating protein 2 |
29357 |
0.16 |
| chr4_103933550_103933744 | 0.49 |
Gm12719 |
predicted gene 12719 |
5894 |
0.25 |
| chr4_135286870_135287040 | 0.49 |
Gm12983 |
predicted gene 12983 |
3720 |
0.15 |
| chr1_171263377_171263582 | 0.49 |
B4galt3 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 3 |
6849 |
0.07 |
| chr11_12411537_12411721 | 0.48 |
Cobl |
cordon-bleu WH2 repeat |
515 |
0.87 |
| chr9_50595278_50595579 | 0.48 |
Sdhd |
succinate dehydrogenase complex, subunit D, integral membrane protein |
8384 |
0.1 |
| chr16_10417919_10418097 | 0.48 |
Nubp1 |
nucleotide binding protein 1 |
3228 |
0.2 |
| chr7_4815487_4815639 | 0.47 |
Ube2s |
ubiquitin-conjugating enzyme E2S |
2973 |
0.11 |
| chr11_16826830_16827159 | 0.47 |
Egfros |
epidermal growth factor receptor, opposite strand |
3708 |
0.27 |
| chr11_16871782_16871933 | 0.47 |
Egfr |
epidermal growth factor receptor |
6293 |
0.22 |
| chr3_142439924_142440082 | 0.47 |
Pdlim5 |
PDZ and LIM domain 5 |
44307 |
0.13 |
| chr11_16899277_16899448 | 0.46 |
Egfr |
epidermal growth factor receptor |
5823 |
0.21 |
| chr15_58983651_58983950 | 0.46 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
336 |
0.87 |
| chr18_15407944_15408276 | 0.45 |
Aqp4 |
aquaporin 4 |
22 |
0.98 |
| chr15_7189723_7189941 | 0.44 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
33237 |
0.19 |
| chr1_170997840_170997991 | 0.44 |
Gm10522 |
predicted gene 10522 |
15971 |
0.08 |
| chr3_122120313_122120499 | 0.44 |
Abca4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
366 |
0.85 |
| chrX_74244741_74244892 | 0.44 |
Flna |
filamin, alpha |
1548 |
0.18 |
| chr4_63402725_63402887 | 0.44 |
Akna |
AT-hook transcription factor |
548 |
0.68 |
| chr10_12821465_12821645 | 0.43 |
Utrn |
utrophin |
5383 |
0.21 |
| chr1_15610696_15611021 | 0.43 |
Gm37138 |
predicted gene, 37138 |
54609 |
0.13 |
| chr11_101060306_101060475 | 0.43 |
Naglu |
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
9622 |
0.09 |
| chr4_43558706_43558873 | 0.43 |
Tln1 |
talin 1 |
329 |
0.72 |
| chr8_11272313_11272503 | 0.43 |
Col4a1 |
collagen, type IV, alpha 1 |
7248 |
0.18 |
| chr4_55485084_55485235 | 0.43 |
Gm22150 |
predicted gene, 22150 |
16000 |
0.13 |
| chr12_81188687_81188872 | 0.43 |
Mir3067 |
microRNA 3067 |
22628 |
0.18 |
| chr11_98775529_98775744 | 0.42 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
303 |
0.81 |
| chr8_121968047_121968241 | 0.42 |
Gm17709 |
predicted gene, 17709 |
5678 |
0.11 |
| chr10_99154735_99154910 | 0.42 |
Poc1b |
POC1 centriolar protein B |
16462 |
0.12 |
| chr16_95728927_95729113 | 0.42 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
11286 |
0.18 |
| chr3_138231897_138232387 | 0.42 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
4091 |
0.14 |
| chr3_95985374_95985550 | 0.41 |
Plekho1 |
pleckstrin homology domain containing, family O member 1 |
10206 |
0.11 |
| chr13_90896944_90897260 | 0.41 |
Atp6ap1l |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
1722 |
0.38 |
| chr2_18381810_18381961 | 0.41 |
Dnajc1 |
DnaJ heat shock protein family (Hsp40) member C1 |
10535 |
0.23 |
| chr12_104346091_104346278 | 0.41 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
7698 |
0.12 |
| chr15_55265818_55265991 | 0.41 |
Gm26296 |
predicted gene, 26296 |
3301 |
0.29 |
| chr13_100543031_100543362 | 0.41 |
Ocln |
occludin |
9264 |
0.12 |
| chr1_170607667_170608002 | 0.40 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
17973 |
0.17 |
| chr9_14396183_14396345 | 0.40 |
Gm47319 |
predicted gene, 47319 |
10572 |
0.11 |
| chr15_94676553_94676729 | 0.40 |
Gm25546 |
predicted gene, 25546 |
4745 |
0.27 |
| chr4_119966810_119966961 | 0.39 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
65881 |
0.12 |
| chr1_155109783_155109942 | 0.39 |
Ier5 |
immediate early response 5 |
10226 |
0.15 |
| chr6_71205289_71205700 | 0.39 |
Fabp1 |
fatty acid binding protein 1, liver |
5667 |
0.13 |
| chr14_22964972_22965155 | 0.39 |
Gm47601 |
predicted gene, 47601 |
37137 |
0.22 |
| chr10_116220817_116221022 | 0.39 |
Ptprr |
protein tyrosine phosphatase, receptor type, R |
24523 |
0.18 |
| chr11_114875733_114875884 | 0.39 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
11316 |
0.12 |
| chr12_103658352_103658505 | 0.39 |
Serpina6 |
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
1216 |
0.31 |
| chr7_110539127_110539556 | 0.39 |
Gm10087 |
predicted gene 10087 |
12549 |
0.18 |
| chr3_79565517_79565668 | 0.39 |
Fnip2 |
folliculin interacting protein 2 |
2087 |
0.19 |
| chr13_45857382_45857567 | 0.39 |
Atxn1 |
ataxin 1 |
14814 |
0.25 |
| chr14_31018658_31018887 | 0.39 |
Gnl3 |
guanine nucleotide binding protein-like 3 (nucleolar) |
217 |
0.61 |
| chr15_77842610_77842777 | 0.39 |
Myh9 |
myosin, heavy polypeptide 9, non-muscle |
518 |
0.7 |
| chr14_51883561_51883781 | 0.38 |
Mettl17 |
methyltransferase like 17 |
1171 |
0.27 |
| chr10_23816629_23816780 | 0.38 |
Slc18b1 |
solute carrier family 18, subfamily B, member 1 |
7783 |
0.12 |
| chr1_160036977_160037141 | 0.38 |
Gm37294 |
predicted gene, 37294 |
7272 |
0.17 |
| chrX_136172356_136172537 | 0.38 |
Tceal8 |
transcription elongation factor A (SII)-like 8 |
104 |
0.94 |
| chr10_99285837_99286030 | 0.37 |
Gm48089 |
predicted gene, 48089 |
11137 |
0.12 |
| chr11_87749523_87749805 | 0.37 |
Mir142hg |
Mir142 host gene (non-protein coding) |
5913 |
0.09 |
| chr14_20081120_20081302 | 0.37 |
Saysd1 |
SAYSVFN motif domain containing 1 |
1972 |
0.31 |
| chr9_46215947_46216118 | 0.37 |
Sik3 |
SIK family kinase 3 |
3539 |
0.13 |
| chr18_7620800_7620994 | 0.37 |
Mpp7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
5920 |
0.24 |
| chr3_127780482_127780684 | 0.37 |
Alpk1 |
alpha-kinase 1 |
56 |
0.96 |
| chr1_33757706_33757887 | 0.37 |
Bag2 |
BCL2-associated athanogene 2 |
1 |
0.97 |
| chr19_53554531_53554692 | 0.37 |
Gm50394 |
predicted gene, 50394 |
24956 |
0.11 |
| chr12_86833864_86834026 | 0.37 |
Gm10095 |
predicted gene 10095 |
12522 |
0.19 |
| chr17_80677489_80677679 | 0.36 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
26434 |
0.19 |
| chr9_74894770_74895613 | 0.36 |
Onecut1 |
one cut domain, family member 1 |
28707 |
0.13 |
| chr4_101490584_101490753 | 0.36 |
Dnajc6 |
DnaJ heat shock protein family (Hsp40) member C6 |
5963 |
0.21 |
| chr5_143055727_143055894 | 0.36 |
Gm43378 |
predicted gene 43378 |
5767 |
0.15 |
| chr18_5186924_5187075 | 0.36 |
Gm26682 |
predicted gene, 26682 |
21268 |
0.24 |
| chr1_63178308_63178488 | 0.36 |
Gm26457 |
predicted gene, 26457 |
279 |
0.62 |
| chr13_56648096_56648259 | 0.36 |
Tgfbi |
transforming growth factor, beta induced |
16964 |
0.19 |
| chr6_142459631_142459782 | 0.36 |
Gys2 |
glycogen synthase 2 |
13403 |
0.16 |
| chr12_35681929_35682122 | 0.35 |
9130015A21Rik |
RIKEN cDNA 9130015A21 gene |
4279 |
0.24 |
| chr7_19131416_19131589 | 0.35 |
Fbxo46 |
F-box protein 46 |
191 |
0.86 |
| chr9_66157716_66157867 | 0.35 |
Dapk2 |
death-associated protein kinase 2 |
432 |
0.83 |
| chr5_129415440_129415591 | 0.35 |
Gm43001 |
predicted gene 43001 |
51395 |
0.13 |
| chr13_4771823_4771989 | 0.35 |
Gm5444 |
predicted gene 5444 |
255 |
0.94 |
| chr19_30246847_30246998 | 0.35 |
Mbl2 |
mannose-binding lectin (protein C) 2 |
13980 |
0.19 |
| chr6_128662223_128662572 | 0.35 |
Clec2h |
C-type lectin domain family 2, member h |
6 |
0.89 |
| chr3_56014019_56014179 | 0.35 |
Gm42840 |
predicted gene 42840 |
3069 |
0.25 |
| chr11_16885540_16885691 | 0.35 |
Egfr |
epidermal growth factor receptor |
7465 |
0.21 |
| chr13_28654991_28655297 | 0.35 |
Mir6368 |
microRNA 6368 |
55729 |
0.15 |
| chr17_25082238_25082418 | 0.35 |
Tmem204 |
transmembrane protein 204 |
1138 |
0.34 |
| chr6_90603208_90603366 | 0.35 |
Slc41a3 |
solute carrier family 41, member 3 |
1438 |
0.31 |
| chr15_7194778_7194945 | 0.34 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
28208 |
0.2 |
| chr10_18069023_18069192 | 0.34 |
Reps1 |
RalBP1 associated Eps domain containing protein |
13103 |
0.19 |
| chr9_107651281_107651577 | 0.34 |
Slc38a3 |
solute carrier family 38, member 3 |
4591 |
0.08 |
| chr6_99364656_99364808 | 0.34 |
Gm20705 |
predicted gene 20705 |
23767 |
0.22 |
| chr9_21167812_21167963 | 0.34 |
Pde4a |
phosphodiesterase 4A, cAMP specific |
2173 |
0.17 |
| chr7_127819457_127819621 | 0.33 |
Stx4a |
syntaxin 4A (placental) |
4755 |
0.08 |
| chr9_7959424_7959575 | 0.33 |
Yap1 |
yes-associated protein 1 |
6322 |
0.18 |
| chr9_50492228_50492379 | 0.33 |
Plet1 |
placenta expressed transcript 1 |
2222 |
0.22 |
| chr7_130518318_130518504 | 0.33 |
Ate1 |
arginyltransferase 1 |
1050 |
0.41 |
| chr12_69517687_69517891 | 0.33 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
42239 |
0.1 |
| chr11_51061072_51061284 | 0.33 |
Zfp354a |
zinc finger protein 354A |
1056 |
0.35 |
| chr12_112720973_112721159 | 0.33 |
Cep170b |
centrosomal protein 170B |
611 |
0.56 |
| chr9_95194235_95194412 | 0.33 |
Slc9a9 |
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
72637 |
0.11 |
| chr2_37578235_37578422 | 0.33 |
Strbp |
spermatid perinuclear RNA binding protein |
8190 |
0.18 |
| chr12_41084537_41084707 | 0.33 |
Immp2l |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
16310 |
0.21 |
| chr15_12209919_12210084 | 0.32 |
Mtmr12 |
myotubularin related protein 12 |
4883 |
0.13 |
| chr3_62349486_62349833 | 0.32 |
Arhgef26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
8558 |
0.24 |
| chr8_34951587_34951746 | 0.32 |
Gm9939 |
predicted gene 9939 |
13386 |
0.17 |
| chr15_85042397_85042828 | 0.32 |
Fam118a |
family with sequence similarity 118, member A |
130 |
0.94 |
| chrX_99292874_99293036 | 0.32 |
Gm14809 |
predicted gene 14809 |
125940 |
0.05 |
| chr9_9104761_9104983 | 0.32 |
Gm16833 |
predicted gene, 16833 |
131416 |
0.05 |
| chr17_30458453_30458632 | 0.32 |
Gm50250 |
predicted gene, 50250 |
8144 |
0.16 |
| chr13_23556410_23556600 | 0.31 |
H1f3 |
H1.3 linker histone, cluster member |
3243 |
0.06 |
| chr14_22493467_22493618 | 0.31 |
4930405A10Rik |
RIKEN cDNA 4930405A10 gene |
49256 |
0.15 |
| chr6_23098041_23098398 | 0.31 |
Aass |
aminoadipate-semialdehyde synthase |
29361 |
0.14 |
| chr17_81006523_81006701 | 0.31 |
Thumpd2 |
THUMP domain containing 2 |
58455 |
0.12 |
| chr8_111828693_111828997 | 0.31 |
Cfdp1 |
craniofacial development protein 1 |
25446 |
0.14 |
| chr12_72482174_72482506 | 0.31 |
Lrrc9 |
leucine rich repeat containing 9 |
6266 |
0.2 |
| chr13_41643806_41643994 | 0.31 |
Gm5082 |
predicted gene 5082 |
5543 |
0.21 |
| chr11_106412539_106412732 | 0.31 |
Icam2 |
intercellular adhesion molecule 2 |
24560 |
0.13 |
| chr1_92724078_92724229 | 0.31 |
Gm29483 |
predicted gene 29483 |
6951 |
0.15 |
| chr3_119042748_119042912 | 0.31 |
Gm43410 |
predicted gene 43410 |
174586 |
0.03 |
| chr19_4134482_4134777 | 0.31 |
Cabp4 |
calcium binding protein 4 |
4697 |
0.06 |
| chr12_41345678_41346003 | 0.31 |
Gm47376 |
predicted gene, 47376 |
2250 |
0.28 |
| chr2_58773576_58773851 | 0.31 |
Upp2 |
uridine phosphorylase 2 |
8388 |
0.21 |
| chr13_28466271_28466432 | 0.31 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
14490 |
0.21 |
| chr1_170006602_170006897 | 0.31 |
3110045C21Rik |
RIKEN cDNA 3110045C21 gene |
37297 |
0.13 |
| chr2_83714821_83715007 | 0.31 |
Itgav |
integrin alpha V |
9483 |
0.16 |
| chr1_130387327_130387514 | 0.31 |
Cd55b |
CD55 molecule, decay accelerating factor for complement B |
35320 |
0.11 |
| chr11_16911020_16911171 | 0.31 |
Egfr |
epidermal growth factor receptor |
5910 |
0.2 |
| chr7_89579648_89579808 | 0.30 |
Gm45064 |
predicted gene 45064 |
27362 |
0.13 |
| chr3_121340564_121340760 | 0.30 |
Gm5711 |
predicted gene 5711 |
40017 |
0.11 |
| chr4_95207881_95208040 | 0.30 |
Gm12708 |
predicted gene 12708 |
14598 |
0.2 |
| chr3_129767244_129767395 | 0.30 |
Egf |
epidermal growth factor |
12003 |
0.12 |
| chr14_66870804_66870955 | 0.30 |
Gm5464 |
predicted gene 5464 |
2029 |
0.24 |
| chr7_141429982_141430161 | 0.30 |
Cend1 |
cell cycle exit and neuronal differentiation 1 |
580 |
0.43 |
| chr15_66783892_66784052 | 0.30 |
Tg |
thyroglobulin |
14778 |
0.18 |
| chr3_142569357_142569508 | 0.30 |
Gbp3 |
guanylate binding protein 3 |
8198 |
0.13 |
| chr10_79838143_79838304 | 0.30 |
Misp |
mitotic spindle positioning |
8937 |
0.06 |
| chr1_91104292_91104494 | 0.30 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
6821 |
0.19 |
| chr7_27357399_27357576 | 0.30 |
Gm20479 |
predicted gene 20479 |
68 |
0.94 |
| chr1_4544211_4544388 | 0.30 |
Gm38076 |
predicted gene, 38076 |
9013 |
0.13 |
| chr2_53102207_53102377 | 0.30 |
Prpf40a |
pre-mRNA processing factor 40A |
43017 |
0.19 |
| chr17_13441059_13441220 | 0.30 |
Rnu6 |
U6 small nuclear RNA |
9000 |
0.12 |
| chr13_98952126_98952306 | 0.30 |
Gm35215 |
predicted gene, 35215 |
6300 |
0.13 |
| chr7_65737343_65737722 | 0.30 |
Tm2d3 |
TM2 domain containing 3 |
44008 |
0.12 |
| chr10_41992198_41992354 | 0.30 |
Armc2 |
armadillo repeat containing 2 |
890 |
0.62 |
| chr6_34722780_34722959 | 0.29 |
Npn2 |
neoplastic progression 2 |
4036 |
0.16 |
| chr10_95408627_95408789 | 0.29 |
Socs2 |
suppressor of cytokine signaling 2 |
4026 |
0.16 |
| chr16_35075564_35075912 | 0.29 |
Hacd2 |
3-hydroxyacyl-CoA dehydratase 2 |
27123 |
0.17 |
| chr15_77816105_77816293 | 0.29 |
Myh9 |
myosin, heavy polypeptide 9, non-muscle |
2968 |
0.2 |
| chr17_13144924_13145092 | 0.29 |
Unc93a |
unc-93 homolog A |
13198 |
0.12 |
| chr5_28079367_28079544 | 0.29 |
Insig1 |
insulin induced gene 1 |
4789 |
0.19 |
| chr6_116653345_116653528 | 0.29 |
Depp1 |
DEPP1 autophagy regulator |
2740 |
0.15 |
| chr19_40478313_40478475 | 0.29 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
9161 |
0.21 |
| chr5_119877302_119877469 | 0.29 |
Gm43269 |
predicted gene 43269 |
37553 |
0.13 |
| chr7_80044210_80044553 | 0.29 |
Gm44951 |
predicted gene 44951 |
7676 |
0.12 |
| chr10_95254224_95254433 | 0.29 |
Gm48880 |
predicted gene, 48880 |
60525 |
0.08 |
| chr4_86318074_86318389 | 0.29 |
Adamtsl1 |
ADAMTS-like 1 |
41465 |
0.19 |
| chr6_31090797_31090948 | 0.29 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
2401 |
0.18 |
| chr5_124232389_124232618 | 0.29 |
Pitpnm2os1 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 1 |
2778 |
0.16 |
| chr11_83271635_83271942 | 0.29 |
Gm23444 |
predicted gene, 23444 |
290 |
0.8 |
| chr10_61160789_61160964 | 0.29 |
Tbata |
thymus, brain and testes associated |
11078 |
0.15 |
| chr6_122310324_122310479 | 0.29 |
M6pr |
mannose-6-phosphate receptor, cation dependent |
1402 |
0.29 |
| chr12_108370797_108370948 | 0.29 |
Eml1 |
echinoderm microtubule associated protein like 1 |
85 |
0.97 |
| chr7_127973702_127973919 | 0.29 |
Fus |
fused in sarcoma |
333 |
0.72 |
| chr5_123382074_123382225 | 0.28 |
Mlxip |
MLX interacting protein |
12649 |
0.07 |
| chr11_69667800_69667951 | 0.28 |
Gm24029 |
predicted gene, 24029 |
101 |
0.56 |
| chr11_59994414_59994592 | 0.28 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
10763 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0072262 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.1 | 0.4 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.3 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.4 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.5 | GO:0072711 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.1 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.4 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.3 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:1900238 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.3 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.6 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.2 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.2 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0001805 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:1902946 | protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0018119 | peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.1 | GO:0060956 | endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1901341 | positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 0.0 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0018904 | ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) |
| 0.0 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.0 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:1901033 | positive regulation of response to reactive oxygen species(GO:1901033) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.0 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0061620 | glycolytic process through glucose-6-phosphate(GO:0061620) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0035729 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.6 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.3 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0034901 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
| 0.0 | 0.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |